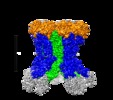
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0264 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Type VI membrane complex | |||||||||
 Map data Map data | MolRes Sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Membrane complex / tether / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.6 Å | |||||||||
 Authors Authors | Rapisarda C / Fronzes R | |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2019 Journal: EMBO J / Year: 2019Title: and high-resolution cryo-EM structure of a bacterial type VI secretion system membrane complex. Authors: Chiara Rapisarda / Yassine Cherrak / Romain Kooger / Victoria Schmidt / Riccardo Pellarin / Laureen Logger / Eric Cascales / Martin Pilhofer / Eric Durand / Rémi Fronzes /   Abstract: Bacteria have evolved macromolecular machineries that secrete effectors and toxins to survive and thrive in diverse environments. The type VI secretion system (T6SS) is a contractile machine that is ...Bacteria have evolved macromolecular machineries that secrete effectors and toxins to survive and thrive in diverse environments. The type VI secretion system (T6SS) is a contractile machine that is related to phages. It is composed of a phage tail-like structure inserted in the bacterial cell envelope by a membrane complex (MC) comprising the TssJ, TssL and TssM proteins. We previously reported the low-resolution negative-stain electron microscopy structure of the enteroaggregative MC and proposed a rotational 5-fold symmetry with a TssJ:TssL:TssM stoichiometry of 2:2:2. Here, cryo-electron tomography analyses of the T6SS MC confirm the 5-fold symmetry and identify the regions of the structure that insert into the bacterial membranes. A high-resolution model obtained by single-particle cryo-electron microscopy highlights new features: five additional copies of TssJ, yielding a TssJ:TssL:TssM stoichiometry of 3:2:2, an 11-residue loop in TssM, protruding inside the lumen of the MC and constituting a functionally important periplasmic gate, and hinge regions. Based on these data, we propose an updated model on MC structure and dynamics during T6SS assembly and function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0264.map.gz emd_0264.map.gz | 372.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0264-v30.xml emd-0264-v30.xml emd-0264.xml emd-0264.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0264.png emd_0264.png | 164.1 KB | ||
| Filedesc metadata |  emd-0264.cif.gz emd-0264.cif.gz | 6.3 KB | ||
| Others |  emd_0264_additional.map.gz emd_0264_additional.map.gz emd_0264_half_map_1.map.gz emd_0264_half_map_1.map.gz emd_0264_half_map_2.map.gz emd_0264_half_map_2.map.gz | 377.2 MB 380.6 MB 381.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0264 http://ftp.pdbj.org/pub/emdb/structures/EMD-0264 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0264 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0264 | HTTPS FTP |
-Validation report
| Summary document |  emd_0264_validation.pdf.gz emd_0264_validation.pdf.gz | 345.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_0264_full_validation.pdf.gz emd_0264_full_validation.pdf.gz | 344.9 KB | Display | |
| Data in XML |  emd_0264_validation.xml.gz emd_0264_validation.xml.gz | 15.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0264 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0264 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0264 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0264 | HTTPS FTP |
-Related structure data
| Related structure data |  6hs7MC  0265C  0266C  0267C  4561C  4562C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0264.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0264.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | MolRes Sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.24 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Refinement map
| File | emd_0264_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refinement map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_0264_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_0264_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Membrane complex of the type VI secretion system (TssM and TssJ)
| Entire | Name: Membrane complex of the type VI secretion system (TssM and TssJ) |
|---|---|
| Components |
|
-Supramolecule #1: Membrane complex of the type VI secretion system (TssM and TssJ)
| Supramolecule | Name: Membrane complex of the type VI secretion system (TssM and TssJ) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: ImcF-like family protein
| Macromolecule | Name: ImcF-like family protein / type: protein_or_peptide / ID: 1 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 127.255367 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNKLACLSGR FGRPGIVFIG VAALWWLITR YGAFLGAETR RDQILLLILL SLGLLFVCYL PVMKKYVQEL TYRRRARKEQ RLPDDEERL AQTPPRYVTV QDIRHTLRRQ YGRFWGRKIR ILLITGTASE VELLTPGLTE QFWQEEQGTL LLWGGEPSQP E NADWLAAL ...String: MNKLACLSGR FGRPGIVFIG VAALWWLITR YGAFLGAETR RDQILLLILL SLGLLFVCYL PVMKKYVQEL TYRRRARKEQ RLPDDEERL AQTPPRYVTV QDIRHTLRRQ YGRFWGRKIR ILLITGTASE VELLTPGLTE QFWQEEQGTL LLWGGEPSQP E NADWLAAL RRLRYRPADG IVWVTSGLSE TLSAPLTEDA LDRVSRAVSS CCERLGWRLP LYVWSLQESP DERGRITQPV GC LLPAECS SDKLKAQLQA MLPGLVAQGI QQICCAPRYY FLLSLAERFR RNIDAVVEPL SVLLRPYRQL LLAGIVFSPA TVG GERSVR HRWRMDNRWE ALPETVQQLP VRLQPSRTGH NWRRSLAVMA AILMMAQGTG MVVSFLANRS LVAEVQEQIR PAQN QQLSP AERLQALLNL QKSLARLQYR EEHGAPWYLR AGMNQNADLL VVVMPLYAQN AHLLLRDAAA AHLEQQLRTF IRLPP DSPQ RGKMAKAAYD QLRLYLMLAQ PQHMEPAWFS RTLMREWPQR DGVSAVFWQA NGPTLLAYYA SGIITHPQWK LTADEE LVS QSRTLLLRHL GTQNSDAMLY QKMLARVAHQ FADMRLTDMT GDTDVSRLFF TDEVVPGMFT RQAWEEAVLP SIDTVIN ER REEMDWVLTD GRQKAPSPVS PEALRQRLTT RYFADFGNAW LNFLNSLHLR KAQMLSDVTE QLTLMADVRQ SPLVALMN T LAVQGRTGQP REAVTDSLVK SARNLLSQEK QPVAVPESRL HGPLATTFGP VLALMDNQNN SADMLNLQTY LTRVTQVRL RLQQIAGSSD PQAMMQMLAQ TVLQGKSVDL TDTRDYGSLT AAGLGQEWYG FGQTVFVRPM EQAWQQVLTP AAESLNARWR TAVVDGWNN AFSGRYPFKN VSSDASLPLL AKYLNTDTGR IARFLQNNLS GVLHREGSRW VPDTINTRGL TFNPAFLKAI N TLSEIADV AFTTGNAGLH FELRPGTAAG VMQTTLITDN QKLIYVNQMP VWKRFTWPAD TEAPGASLSW VSTQAGTRQY AD LPGSWGL IRLLEMARRK AAPGVASGWS LSWQAQDGRM LNYTLRTEAG EGPLVLLKLR NFVLPETVFE LSGTSAFTGN DED AGDTVE ETD UniProtKB: ImcF-like family protein |
-Macromolecule #2: Type VI secretion system protein VasD
| Macromolecule | Name: Type VI secretion system protein VasD / type: protein_or_peptide / ID: 2 / Number of copies: 15 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 20.313236 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAIIAGKAGY GLIIALFSLS LSGCGLTQRV ADGTVSATKS LFYRQIKTLH LDIRAREAIN TSAAGIPLSV VVRIYQLKDN RSFDSADYQ ALFTGDNEIL AGDIIAQKDV WLQPGGSVAV DMPLDDAAKF TGVAAMFLEP DQKKNTWRVV LGRDELEPDT P RLIEVSGN TLTLLPVKDK WSHPQFEK UniProtKB: Type VI secretion system protein VasD |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: C-flat-2/2 / Material: COPPER / Mesh: 200 / Support film - Material: GRAPHENE OXIDE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 120.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 0.003 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 0.002 µm / Nominal defocus min: 0.0005 µm / Nominal magnification: 120000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C5 (5 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 4.6 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION / Number images used: 36828 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: cryoSPARC |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)