+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0241 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Yeast apo RNA polymerase I* | |||||||||
 Map data Map data | Yeast apo RNA polymerase I* | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transcription / polymerase / nucleotide / elongation | |||||||||
| Function / homology |  Function and homology information Function and homology informationRNA Polymerase I Transcription Initiation / Processing of Capped Intron-Containing Pre-mRNA / RNA Polymerase III Transcription Initiation From Type 2 Promoter / RNA Pol II CTD phosphorylation and interaction with CE / Formation of the Early Elongation Complex / mRNA Capping / RNA polymerase II transcribes snRNA genes / TP53 Regulates Transcription of DNA Repair Genes / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening ...RNA Polymerase I Transcription Initiation / Processing of Capped Intron-Containing Pre-mRNA / RNA Polymerase III Transcription Initiation From Type 2 Promoter / RNA Pol II CTD phosphorylation and interaction with CE / Formation of the Early Elongation Complex / mRNA Capping / RNA polymerase II transcribes snRNA genes / TP53 Regulates Transcription of DNA Repair Genes / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance / termination of RNA polymerase III transcription / RNA Polymerase II Pre-transcription Events / RNA-templated transcription / Formation of TC-NER Pre-Incision Complex / regulation of cell size / RNA Polymerase I Promoter Escape / transcription initiation at RNA polymerase III promoter / termination of RNA polymerase I transcription / Gap-filling DNA repair synthesis and ligation in TC-NER / nucleolar large rRNA transcription by RNA polymerase I / transcription initiation at RNA polymerase I promoter / Estrogen-dependent gene expression / transcription by RNA polymerase III / Dual incision in TC-NER / RNA polymerase I complex / transcription elongation by RNA polymerase I / RNA polymerase III complex / tRNA transcription by RNA polymerase III / RNA polymerase II, core complex / transcription by RNA polymerase I / transcription initiation at RNA polymerase II promoter / transcription elongation by RNA polymerase II / promoter-specific chromatin binding / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / peroxisome / ribosome biogenesis / nucleic acid binding / RNA polymerase II-specific DNA-binding transcription factor binding / transcription by RNA polymerase II / protein dimerization activity / nucleolus / negative regulation of transcription by RNA polymerase II / mitochondrion / DNA binding / zinc ion binding / nucleoplasm / metal ion binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
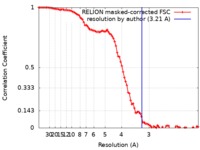
| Method | single particle reconstruction / cryo EM / Resolution: 3.21 Å | |||||||||
 Authors Authors | Tafur L / Sadian Y | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2019 Journal: Elife / Year: 2019Title: The cryo-EM structure of a 12-subunit variant of RNA polymerase I reveals dissociation of the A49-A34.5 heterodimer and rearrangement of subunit A12.2. Authors: Lucas Tafur / Yashar Sadian / Jonas Hanske / Rene Wetzel / Felix Weis / Christoph W Müller /  Abstract: RNA polymerase (Pol) I is a 14-subunit enzyme that solely transcribes pre-ribosomal RNA. Cryo-electron microscopy (EM) structures of Pol I initiation and elongation complexes have given first ...RNA polymerase (Pol) I is a 14-subunit enzyme that solely transcribes pre-ribosomal RNA. Cryo-electron microscopy (EM) structures of Pol I initiation and elongation complexes have given first insights into the molecular mechanisms of Pol I transcription. Here, we present cryo-EM structures of yeast Pol I elongation complexes (ECs) bound to the nucleotide analog GMPCPP at 3.2 to 3.4 Å resolution that provide additional insight into the functional interplay between the Pol I-specific transcription-like factors A49-A34.5 and A12.2. Strikingly, most of the nucleotide-bound ECs lack the A49-A34.5 heterodimer and adopt a Pol II-like conformation, in which the A12.2 C-terminal domain is bound in a previously unobserved position at the A135 surface. Our structural and biochemical data suggest a mechanism where reversible binding of the A49-A34.5 heterodimer could contribute to the regulation of Pol I transcription initiation and elongation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0241.map.gz emd_0241.map.gz | 62.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0241-v30.xml emd-0241-v30.xml emd-0241.xml emd-0241.xml | 25.8 KB 25.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0241_fsc.xml emd_0241_fsc.xml | 9.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_0241.png emd_0241.png | 96.8 KB | ||
| Filedesc metadata |  emd-0241.cif.gz emd-0241.cif.gz | 8.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0241 http://ftp.pdbj.org/pub/emdb/structures/EMD-0241 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0241 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0241 | HTTPS FTP |
-Related structure data
| Related structure data |  6hlsMC  0238C  0239C  0240C  0242C  6hkoC  6hlqC  6hlrC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0241.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0241.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Yeast apo RNA polymerase I* | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Yeast apo RNA polymerase I*
+Supramolecule #1: Yeast apo RNA polymerase I*
+Macromolecule #1: DNA-directed RNA polymerase I subunit RPA190
+Macromolecule #2: DNA-directed RNA polymerase I subunit RPA135
+Macromolecule #3: DNA-directed RNA polymerases I and III subunit RPAC1
+Macromolecule #4: DNA-directed RNA polymerase I subunit RPA14
+Macromolecule #5: DNA-directed RNA polymerases I, II, and III subunit RPABC1
+Macromolecule #6: DNA-directed RNA polymerases I, II, and III subunit RPABC2
+Macromolecule #7: DNA-directed RNA polymerase I subunit RPA43
+Macromolecule #8: DNA-directed RNA polymerases I, II, and III subunit RPABC3
+Macromolecule #9: DNA-directed RNA polymerase I subunit RPA12
+Macromolecule #10: DNA-directed RNA polymerases I, II, and III subunit RPABC5
+Macromolecule #11: DNA-directed RNA polymerases I and III subunit RPAC2
+Macromolecule #12: DNA-directed RNA polymerases I, II, and III subunit RPABC4
+Macromolecule #13: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: 30 seconds incubation 3 seconds blotting blotting force 3. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 39.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Details | Initial rigid body fitting was done in Chimera and model building was done in Coot. | ||||||
| Refinement | Space: REAL / Protocol: RIGID BODY FIT | ||||||
| Output model |  PDB-6hls: |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)