[English] 日本語
 Yorodumi
Yorodumi- EMDB-0219: Cryo-EM structure 2 of pre-60S particles isolated via Arx1-FTpA f... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0219 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure 2 of pre-60S particles isolated via Arx1-FTpA from yeast | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
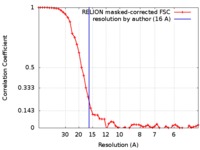
| Method | single particle reconstruction / cryo EM / Resolution: 16.0 Å | |||||||||
 Authors Authors | Kater L / Beckmann R | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: Suppressor mutations in Rpf2-Rrs1 or Rpl5 bypass the Cgr1 function for pre-ribosomal 5S RNP-rotation. Authors: Matthias Thoms / Valentin Mitterer / Lukas Kater / Laurent Falquet / Roland Beckmann / Dieter Kressler / Ed Hurt /   Abstract: During eukaryotic 60S biogenesis, the 5S RNP requires a large rotational movement to achieve its mature position. Cryo-EM of the Rix1-Rea1 pre-60S particle has revealed the post-rotation stage, in ...During eukaryotic 60S biogenesis, the 5S RNP requires a large rotational movement to achieve its mature position. Cryo-EM of the Rix1-Rea1 pre-60S particle has revealed the post-rotation stage, in which a gently undulating α-helix corresponding to Cgr1 becomes wedged between Rsa4 and the relocated 5S RNP, but the purpose of this insertion was unknown. Here, we show that cgr1 deletion in yeast causes a slow-growth phenotype and reversion of the pre-60S particle to the pre-rotation stage. However, spontaneous extragenic suppressors could be isolated, which restore growth and pre-60S biogenesis in the absence of Cgr1. Whole-genome sequencing reveals that the suppressor mutations map in the Rpf2-Rrs1 module and Rpl5, which together stabilize the unrotated stage of the 5S RNP. Thus, mutations in factors stabilizing the pre-rotation stage facilitate 5S RNP relocation upon deletion of Cgr1, but Cgr1 itself could stabilize the post-rotation stage. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0219.map.gz emd_0219.map.gz | 7.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0219-v30.xml emd-0219-v30.xml emd-0219.xml emd-0219.xml | 8.2 KB 8.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0219_fsc.xml emd_0219_fsc.xml | 5.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_0219.png emd_0219.png | 45.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0219 http://ftp.pdbj.org/pub/emdb/structures/EMD-0219 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0219 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0219 | HTTPS FTP |
-Related structure data
| Related structure data |  0218C  0220C  0221C  0222C  0223C  0224C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0219.map.gz / Format: CCP4 / Size: 10.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0219.map.gz / Format: CCP4 / Size: 10.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.55 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Yeast pre60S particle purified via Arx1-FTpA
| Entire | Name: Yeast pre60S particle purified via Arx1-FTpA |
|---|---|
| Components |
|
-Supramolecule #1: Yeast pre60S particle purified via Arx1-FTpA
| Supramolecule | Name: Yeast pre60S particle purified via Arx1-FTpA / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F216 (2k x 2k) / Average electron dose: 32.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)