+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0199 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | human STEAP4 bound to NADP, FAD, heme and Fe(III)-NTA. | ||||||||||||
 Map data Map data | None | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Enzyme / Metalloreductase / Electron Transfer / Cofactor-binding / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationOxidoreductases; Oxidizing metal ions; With NAD+ or NADP+ as acceptor / ferric-chelate reductase (NADPH) activity / cupric reductase (NADH) activity / copper ion import / iron import into cell / Transferrin endocytosis and recycling / protein homotrimerization / fat cell differentiation / FAD binding / early endosome membrane ...Oxidoreductases; Oxidizing metal ions; With NAD+ or NADP+ as acceptor / ferric-chelate reductase (NADPH) activity / cupric reductase (NADH) activity / copper ion import / iron import into cell / Transferrin endocytosis and recycling / protein homotrimerization / fat cell differentiation / FAD binding / early endosome membrane / electron transfer activity / endosome / endosome membrane / Golgi membrane / heme binding / extracellular exosome / nucleoplasm / metal ion binding / membrane / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | ||||||||||||
 Authors Authors | Oosterheert W / van Bezouwen LS | ||||||||||||
| Funding support |  Netherlands, Netherlands,  Italy, 3 items Italy, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: Cryo-EM structures of human STEAP4 reveal mechanism of iron(III) reduction. Authors: Wout Oosterheert / Laura S van Bezouwen / Remco N P Rodenburg / Joke Granneman / Friedrich Förster / Andrea Mattevi / Piet Gros /   Abstract: Enzymes of the six-transmembrane epithelial antigen of the prostate (STEAP) family reduce Fe and Cu ions to facilitate metal-ion uptake by mammalian cells. STEAPs are highly upregulated in several ...Enzymes of the six-transmembrane epithelial antigen of the prostate (STEAP) family reduce Fe and Cu ions to facilitate metal-ion uptake by mammalian cells. STEAPs are highly upregulated in several types of cancer, making them potential therapeutic targets. However, the structural basis for STEAP-catalyzed electron transfer through an array of cofactors to metals at the membrane luminal side remains elusive. Here, we report cryo-electron microscopy structures of human STEAP4 in absence and presence of Fe-NTA. Domain-swapped, trimeric STEAP4 orients NADPH bound to a cytosolic domain onto axially aligned flavin-adenine dinucleotide (FAD) and a single b-type heme that cross the transmembrane-domain to enable electron transfer. Substrate binding within a positively charged ring indicates that iron gets reduced while in complex with its chelator. These molecular principles of iron reduction provide a basis for exploring STEAPs as therapeutic targets. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0199.map.gz emd_0199.map.gz | 14.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0199-v30.xml emd-0199-v30.xml emd-0199.xml emd-0199.xml | 28.6 KB 28.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0199_fsc.xml emd_0199_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_0199.png emd_0199.png | 197.2 KB | ||
| Masks |  emd_0199_msk_1.map emd_0199_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-0199.cif.gz emd-0199.cif.gz | 7.5 KB | ||
| Others |  emd_0199_additional_1.map.gz emd_0199_additional_1.map.gz emd_0199_additional_2.map.gz emd_0199_additional_2.map.gz emd_0199_additional_3.map.gz emd_0199_additional_3.map.gz emd_0199_half_map_1.map.gz emd_0199_half_map_1.map.gz emd_0199_half_map_2.map.gz emd_0199_half_map_2.map.gz | 80.3 MB 94.5 MB 94.9 MB 80.7 MB 80.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0199 http://ftp.pdbj.org/pub/emdb/structures/EMD-0199 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0199 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0199 | HTTPS FTP |
-Related structure data
| Related structure data |  6hcyMC  0200C  6hd1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0199.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0199.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8127 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
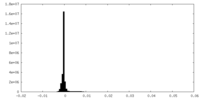
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_0199_msk_1.map emd_0199_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
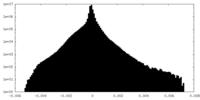
| Density Histograms |
-Additional map: Unsharpened, unmasked density map of human STEAP4 in...
| File | emd_0199_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened, unmasked density map of human STEAP4 in the presence of NADP, FAD, heme and Fe(III)-NTA (cofactor/substrate-bound state). Generated by 3D auto-refine in Relion2.1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened, density map of human STEAP4 in cofactor/substrate-bound...
| File | emd_0199_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened, density map of human STEAP4 in cofactor/substrate-bound state, with its power spectrum adjusted to be the same as for the lower resolution cofactor-bound map, generated by Relion. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Difference map between STEAP in the absence and...
| File | emd_0199_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Difference map between STEAP in the absence and presence of substrate Fe(III)-NTA. Generated through map subtraction in Relion. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: The first unfiltered-half map of the final refinement...
| File | emd_0199_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The first unfiltered-half map of the final refinement of human STEAP4 in the presence of NADP, FAD, heme and Fe(III)-NTA (cofactor/substrate-bound state). Generated by 3D auto-refine in Relion2.1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: The second unfiltered-half map of the final refinement...
| File | emd_0199_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The second unfiltered-half map of the final refinement of human STEAP4 in the presence of NADP, FAD, heme and Fe(III)-NTA (cofactor/substrate-bound state). Generated by 3D auto-refine in Relion2.1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human Six-Transmembrane Epithelial Antigen of the Prostate 4
| Entire | Name: Human Six-Transmembrane Epithelial Antigen of the Prostate 4 |
|---|---|
| Components |
|
-Supramolecule #1: Human Six-Transmembrane Epithelial Antigen of the Prostate 4
| Supramolecule | Name: Human Six-Transmembrane Epithelial Antigen of the Prostate 4 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 152 KDa |
-Macromolecule #1: Metalloreductase STEAP4
| Macromolecule | Name: Metalloreductase STEAP4 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO EC number: Oxidoreductases; Oxidizing metal ions; With NAD+ or NADP+ as acceptor |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 52.036 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEKTCIDALP LTMNSSEKQE TVCIFGTGDF GRSLGLKMLQ CGYSVVFGSR NPQKTTLLPS GAEVLSYSEA AKKSGIIIIA IHREHYDFL TELTEVLNGK ILVDISNNLK INQYPESNAE YLAHLVPGAH VVKAFNTISA WALQSGALDA SRQVFVCGND S KAKQRVMD ...String: MEKTCIDALP LTMNSSEKQE TVCIFGTGDF GRSLGLKMLQ CGYSVVFGSR NPQKTTLLPS GAEVLSYSEA AKKSGIIIIA IHREHYDFL TELTEVLNGK ILVDISNNLK INQYPESNAE YLAHLVPGAH VVKAFNTISA WALQSGALDA SRQVFVCGND S KAKQRVMD IVRNLGLTPM DQGSLMAAKE IEKYPLQLFP MWRFPFYLSA VLCVFLFFYC VIRDVIYPYV YEKKDNTFRM AI SIPNRIF PITALTLLAL VYLPGVIAAI LQLYRGTKYR RFPDWLDHWM LCRKQLGLVA LGFAFLHVLY TLVIPIRYYV RWR LGNLTV TQAILKKENP FSTSSAWLSD SYVALGILGF FLFVLLGITS LPSVSNAVNW REFRFVQSKL GYLTLILCTA HTLV YGGKR FLSPSNLRWY LPAAYVLGLI IPCTVLVIKF VLIMPCVDNT LTRIRQGWER NSKH UniProtKB: Metalloreductase STEAP4 |
-Macromolecule #2: NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE
| Macromolecule | Name: NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE / type: ligand / ID: 2 / Number of copies: 3 / Formula: NAP |
|---|---|
| Molecular weight | Theoretical: 743.405 Da |
| Chemical component information |  ChemComp-NAP: |
-Macromolecule #3: PROTOPORPHYRIN IX CONTAINING FE
| Macromolecule | Name: PROTOPORPHYRIN IX CONTAINING FE / type: ligand / ID: 3 / Number of copies: 3 / Formula: HEM |
|---|---|
| Molecular weight | Theoretical: 616.487 Da |
| Chemical component information |  ChemComp-HEM: |
-Macromolecule #4: FLAVIN-ADENINE DINUCLEOTIDE
| Macromolecule | Name: FLAVIN-ADENINE DINUCLEOTIDE / type: ligand / ID: 4 / Number of copies: 3 / Formula: FAD |
|---|---|
| Molecular weight | Theoretical: 785.55 Da |
| Chemical component information |  ChemComp-FAD: |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #6: (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate
| Macromolecule | Name: (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate / type: ligand / ID: 6 / Number of copies: 3 / Formula: 44E |
|---|---|
| Molecular weight | Theoretical: 368.36 Da |
| Chemical component information | 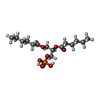 ChemComp-44E: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 5.5 Component:
Details: 25 mM MES pH 5.5 200 mM NaCl 0.08% (w/v) digitonin | ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV / Details: Blotted for 4 seconds Blotforce 0. | ||||||||||||
| Details | The sample was purified from the HEK293 GNTI- cell membrane using STREP-affinity chromatography and size-exclusion chromatography (SEC). The sample was monodisperse after SEC in digitonin. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Details | Grid was prescreened on a 200 kV Talos Arctica Microscope. Microscope alignment was performed by Wim Hagen at EMBL Heidelberg. |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 8211 / Average exposure time: 12.0 sec. / Average electron dose: 47.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | No Fitting was performed. |
|---|---|
| Output model |  PDB-6hcy: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)