+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0151 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rhesus macaque mitochondrial complex I | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
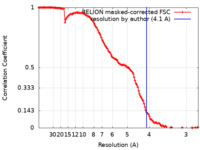
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Agip ANA / Blaza JN / Fedor JG / Hirst J | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Annu Rev Biophys / Year: 2019 Journal: Annu Rev Biophys / Year: 2019Title: Mammalian Respiratory Complex I Through the Lens of Cryo-EM. Authors: Ahmed-Noor A Agip / James N Blaza / Justin G Fedor / Judy Hirst /  Abstract: Single-particle electron cryomicroscopy (cryo-EM) has led to a revolution in structural work on mammalian respiratory complex I. Complex I (mitochondrial NADH:ubiquinone oxidoreductase), a membrane- ...Single-particle electron cryomicroscopy (cryo-EM) has led to a revolution in structural work on mammalian respiratory complex I. Complex I (mitochondrial NADH:ubiquinone oxidoreductase), a membrane-bound redox-driven proton pump, is one of the largest and most complicated enzymes in the mammalian cell. Rapid progress, following the first 5-Å resolution data on bovine complex I in 2014, has led to a model for mouse complex I at 3.3-Å resolution that contains 96% of the 8,518 residues and to the identification of different particle classes, some of which are assigned to biochemically defined states. Factors that helped improve resolution, including improvements to biochemistry, cryo-EM grid preparation, data collection strategy, and image processing, are discussed. Together with recent structural data from an ancient relative, membrane-bound hydrogenase, cryo-EM on mammalian complex I has provided new insights into the proton-pumping machinery and a foundation for understanding the enzyme's catalytic mechanism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0151.map.gz emd_0151.map.gz | 167 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0151-v30.xml emd-0151-v30.xml emd-0151.xml emd-0151.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0151_fsc.xml emd_0151_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_0151.png emd_0151.png | 39.3 KB | ||
| Masks |  emd_0151_msk_1.map emd_0151_msk_1.map | 178 MB |  Mask map Mask map | |
| Others |  emd_0151_half_map_1.map.gz emd_0151_half_map_1.map.gz emd_0151_half_map_2.map.gz emd_0151_half_map_2.map.gz | 140.7 MB 141 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0151 http://ftp.pdbj.org/pub/emdb/structures/EMD-0151 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0151 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0151 | HTTPS FTP |
-Validation report
| Summary document |  emd_0151_validation.pdf.gz emd_0151_validation.pdf.gz | 439 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_0151_full_validation.pdf.gz emd_0151_full_validation.pdf.gz | 438.1 KB | Display | |
| Data in XML |  emd_0151_validation.xml.gz emd_0151_validation.xml.gz | 17.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0151 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0151 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0151 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0151 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0151.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0151.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.39 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_0151_msk_1.map emd_0151_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_0151_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_0151_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Rhesus macaque mitochondrial complex I
| Entire | Name: Rhesus macaque mitochondrial complex I |
|---|---|
| Components |
|
-Supramolecule #1: Rhesus macaque mitochondrial complex I
| Supramolecule | Name: Rhesus macaque mitochondrial complex I / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.14 Component:
| ||||||||||||
| Grid | Model: Quantifoil, UltrAuFoil / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE Details: UltrAuFoil gold grids (0.6/1) was used for the preparation of macaque complex I grids. In brief, UltrAuFoil gold grids (0.6/1) were glow discharged at 20 mA for 60 sec using a EMITECH glow- ...Details: UltrAuFoil gold grids (0.6/1) was used for the preparation of macaque complex I grids. In brief, UltrAuFoil gold grids (0.6/1) were glow discharged at 20 mA for 60 sec using a EMITECH glow-discharger unit. Then, the grids were incubated in 5 mM 11-mercaptoundecyl hexaethyleneglycol (SPT-0011P6, SensoPath Technologies) in ethanol for two days under anaerobic conditions. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 8-16s blotting.. | ||||||||||||
| Details | The sample was collected from the peak fraction of a size-exclusion column and was not concentrated. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Average exposure time: 2.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)