+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Saccharomyces cerevisiae supercomplex class2 (III2IV2) | |||||||||
 Map data Map data | Saccharomyces cerevisiae supercomplex class2 (III2IV2) | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
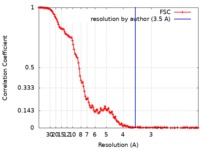
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Rathore S / Berndtsson J / Conrad J / Ott M | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2019 Journal: Nat Struct Mol Biol / Year: 2019Title: Cryo-EM structure of the yeast respiratory supercomplex. Authors: Sorbhi Rathore / Jens Berndtsson / Lorena Marin-Buera / Julian Conrad / Marta Carroni / Peter Brzezinski / Martin Ott /  Abstract: Respiratory chain complexes execute energy conversion by connecting electron transport with proton translocation over the inner mitochondrial membrane to fuel ATP synthesis. Notably, these complexes ...Respiratory chain complexes execute energy conversion by connecting electron transport with proton translocation over the inner mitochondrial membrane to fuel ATP synthesis. Notably, these complexes form multi-enzyme assemblies known as respiratory supercomplexes. Here we used single-particle cryo-EM to determine the structures of the yeast mitochondrial respiratory supercomplexes IIIIV and IIIIV, at 3.2-Å and 3.5-Å resolutions, respectively. We revealed the overall architecture of the supercomplex, which deviates from the previously determined assemblies in mammals; obtained a near-atomic structure of the yeast complex IV; and identified the protein-protein and protein-lipid interactions implicated in supercomplex formation. Take together, our results demonstrate convergent evolution of supercomplexes in mitochondria that, while building similar assemblies, results in substantially different arrangements and structural solutions to support energy conversion. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0006.map.gz emd_0006.map.gz | 149.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0006-v30.xml emd-0006-v30.xml emd-0006.xml emd-0006.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0006_fsc.xml emd_0006_fsc.xml | 15.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_0006.png emd_0006.png | 305.3 KB | ||
| Others |  emd_0006_half_map_1.map.gz emd_0006_half_map_1.map.gz emd_0006_half_map_2.map.gz emd_0006_half_map_2.map.gz | 134 MB 134 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0006 http://ftp.pdbj.org/pub/emdb/structures/EMD-0006 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0006 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0006 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0006.map.gz / Format: CCP4 / Size: 193.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0006.map.gz / Format: CCP4 / Size: 193.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Saccharomyces cerevisiae supercomplex class2 (III2IV2) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
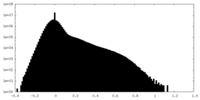
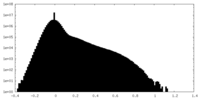
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Halfmap1 Class2
| File | emd_0006_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Halfmap1 Class2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
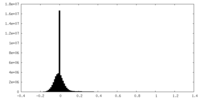
| Density Histograms |
-Half map: Halfmap2 Class2
| File | emd_0006_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Halfmap2 Class2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
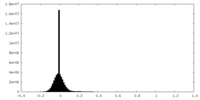
| Density Histograms |
- Sample components
Sample components
-Entire : Saccharomyces cerevisiae Supercomplex
| Entire | Name: Saccharomyces cerevisiae Supercomplex |
|---|---|
| Components |
|
-Supramolecule #1: Saccharomyces cerevisiae Supercomplex
| Supramolecule | Name: Saccharomyces cerevisiae Supercomplex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#25 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)