+ Open data
Open data
- Basic information
Basic information
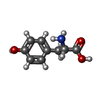
| Entry | 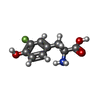 Database: PDB chemical components / ID: YOF Database: PDB chemical components / ID: YOF |
|---|---|
| Name | Name: |
-Chemical information
| Composition |  | ||||||
|---|---|---|---|---|---|---|---|
| Others | Type: L-PEPTIDE LINKING / PDB classification: HETAIN / One letter code: Y / Three letter code: YOF / Model coordinates PDB-ID: 3FYG / Parent comp.: TYR | ||||||
| History |
| ||||||
 External links External links |  UniChem / UniChem /  ChemSpider / ChemSpider /  Wikipedia search / Wikipedia search /  Google search Google search |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
-Details
-SMILES
| ACDLabs 10.04 | | CACTVS 3.341 | OpenEye OEToolkits 1.5.0 | |
|---|
-SMILES CANONICAL
| CACTVS 3.341 | | OpenEye OEToolkits 1.5.0 | |
|---|
-InChI
| InChI 1.03 |
|---|
-InChIKey
| InChI 1.03 |
|---|
-SYSTEMATIC NAME
| ACDLabs 10.04 | | OpenEye OEToolkits 1.5.0 | ( | |
|---|
-PDB entries
Showing all 6 items

PDB-5yak: 
The crystal structure of human IYD Thr239 mutant with ligand 3-Fluorotyrosine (F-Tyr)

PDB-6mpd: 
Citrobacter freundii tyrosine phenol-lyase complexed with 4-hydroxypyridine and aminoacrylate from 3-F-L-tyrosine

PDB-6q1b: 
Crystal structure of oxidized iodotyrosine deiodinase (IYD) bound to FMN and 3-fluoro-L-tyrosine

PDB-7kqs: 
A 1.68-A resolution 3-fluoro-L-tyrosine bound crystal structure of heme-dependent tyrosine hydroxylase

PDB-7kqt: 
A 1.84-A resolution crystal structure of heme-dependent L-tyrosine hydroxylase in complex with 3-fluoro-L-tyrosine and cyanide

PDB-7kqu: 
A 1.58-A resolution crystal structure of ferric-hydroperoxo intermediate of L-tyrosine hydroxylase in complex with 3-fluoro-L-tyrosine
 Movie
Movie Controller
Controller