[English] 日本語
 Yorodumi
Yorodumi- SASDE52: Ribonuclease E from Escherichia coli (Endoribonuclease E, RNase E) -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDE52 |
|---|---|
 Sample Sample | Ribonuclease E from Escherichia coli
|
| Function / homology |  Function and homology information Function and homology informationregulation of RNA helicase activity / rRNA 5'-end processing / ribonuclease E / ribonuclease E activity / bacterial degradosome / endoribonuclease complex / DEAD/H-box RNA helicase binding / 7S RNA binding / RNA catabolic process / tRNA processing ...regulation of RNA helicase activity / rRNA 5'-end processing / ribonuclease E / ribonuclease E activity / bacterial degradosome / endoribonuclease complex / DEAD/H-box RNA helicase binding / 7S RNA binding / RNA catabolic process / tRNA processing / mRNA catabolic process / protein complex oligomerization / RNA nuclease activity / RNA processing / RNA endonuclease activity / cytoplasmic side of plasma membrane / rRNA processing / protein homotetramerization / tRNA binding / molecular adaptor activity / rRNA binding / magnesium ion binding / RNA binding / zinc ion binding / identical protein binding / membrane / cytoplasm Similarity search - Function |
| Biological species |  |
 Citation Citation |  Date: 2019 Dec Date: 2019 DecTitle: A structural and biochemical comparison of Ribonuclease E homologues from pathogenic bacteria highlights species-specific properties Authors: Mardle C / Shakespeare T / Butt L / Goddard L / Gowers D / Atkins H / Vincent H |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDE52 SASDE52 |
|---|
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
| Model #2185 |  Type: dummy / Software: (5) / Radius of dummy atoms: 5.00 A / Symmetry: P222 / Chi-square value: 1.054 / P-value: 0.000144  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|
- Sample
Sample
 Sample Sample | Name: Ribonuclease E from Escherichia coli / Specimen concentration: 6.58 mg/ml |
|---|---|
| Buffer | Name: 10 mM DTT, 10 mM MgCl2, 0.5 M NaCl, 20 mM Tris / pH: 8 |
| Entity #1203 | Name: RNase E / Type: protein / Description: Endoribonuclease E / Formula weight: 61.84 / Num. of mol.: 4 / Source: Escherichia coli / References: UniProt: P21513 Sequence: HHHHHHHHHH SSGHIEGRHM KRMLINATQQ EELRVALVDG QRLYDLDIES PGHEQKKANI YKGKITRIEP SLEAAFVDYG AERHGFLPLK EIAREYFPAN YSAHGRPNIK DVLREGQEVI VQIDKEERGN KGAALTTFIS LAGSYLVLMP NNPRAGGISR RIEGDDRTEL ...Sequence: HHHHHHHHHH SSGHIEGRHM KRMLINATQQ EELRVALVDG QRLYDLDIES PGHEQKKANI YKGKITRIEP SLEAAFVDYG AERHGFLPLK EIAREYFPAN YSAHGRPNIK DVLREGQEVI VQIDKEERGN KGAALTTFIS LAGSYLVLMP NNPRAGGISR RIEGDDRTEL KEALASLELP EGMGLIVRTA GVGKSAEALQ WDLSFRLKHW EAIKKAAESR PAPFLIHQES NVIVRAFRDY LRQDIGEILI DNPKVLELAR QHIAALGRPD FSSKIKLYTG EIPLFSHYQI ESQIESAFQR EVRLPSGGSI VIDSTEALTA IDINSARATR GGDIEETAFN TNLEAADEIA RQLRLRDLGG LIVIDFIDMT PVRHQRAVEN RLREAVRQDR ARIQISHISR FGLLEMSRQR LSPSLGESSH HVCPRCSGTG TVRDNESLSL SILRLIEEEA LKENTQEVHA IVPVPIASYL LNEKRSAVNA IETRQDGVRC VIVPNDQMET PHYHVLRVRK GEETPTLSYM LPKLHEEAMA LPSEEEFAER KRPEQPAL |
-Experimental information
| Beam | Instrument name: Diamond Light Source B21 / City: Oxfordshire / 国: UK  / Shape: 1 x 5 mm / Type of source: X-ray synchrotron / Wavelength: 0.1 Å / Dist. spec. to detc.: 4.014 mm / Shape: 1 x 5 mm / Type of source: X-ray synchrotron / Wavelength: 0.1 Å / Dist. spec. to detc.: 4.014 mm | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 2M | ||||||||||||||||||||||||||||||
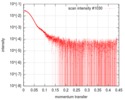
| Scan |
| ||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| ||||||||||||||||||||||||||||||
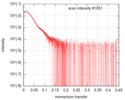
| Result |
|
 Movie
Movie Controller
Controller