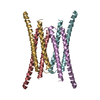[English] 日本語
 Yorodumi
Yorodumi- SASDDN8: Integrin beta4, fragment of the cytoplasmic region that includes ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDDN8 |
|---|---|
 Sample Sample | Integrin beta4, fragment of the cytoplasmic region that includes the final part of the connecting segment and the third and fourth FnIII domains (FnIII-3,4). Point mutant R1542A
|
| Function / homology | Isoform Beta-4A of Integrin beta-4 Function and homology information Function and homology information |
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
 Citation Citation |  Date: 2018 Aug 31 Date: 2018 Aug 31Title: Integrin α6β4 recognition of a linear motif of bullous pemphigoid antigen BP230 controls its recruitment to hemidesmosomes: Supplemental figures S1 to S10 and table S1 Authors: Manso J / Gómez-Hernández M / Carabias A / Alonso-García N / García-Rubio I / Kreft M / Sonnenberg A |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Models
| Model #2068 |  Type: dummy / Software: (r9988) / Radius of dummy atoms: 1.50 A / Chi-square value: 1.047 / P-value: 0.237400  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|
- Sample
Sample
 Sample Sample | Name: Integrin beta4, fragment of the cytoplasmic region that includes the final part of the connecting segment and the third and fourth FnIII domains (FnIII-3,4). Point mutant R1542A Specimen concentration: 0.90-14.50 |
|---|---|
| Buffer | Name: 20 mM Sodium Phosphate 150 mM NaCl 5% glycerol 3 mM DTT Concentration: 20.00 mM / pH: 7.5 / Composition: 150 mM NaCl, 5% glycerol, 3 mM DTT |
| Entity #1107 | Name: B4 CS-FnIII-3,4 / Type: protein / Description: Integrin beta-4 (1436-1666) R1542A / Formula weight: 25.52 / Num. of mol.: 1 / Source: Homo sapiens / References: UniProt: P16144-2 Sequence: GSHMLPRDYS TLTSVSSHDS RLTAGVPDTP TRLVFSALGP TSLRVSWQEP RCERPLQGYS VEYQLLNGGE LHRLNIPNPA QTSVVVEDLL PNHSYVFRVR AQSQEGWGRE AEGVITIESQ VHPQSPLCPL PGSAFTLSTP SAPGPLVFTA LSPDSLQLSW ERPRRPNGDI ...Sequence: GSHMLPRDYS TLTSVSSHDS RLTAGVPDTP TRLVFSALGP TSLRVSWQEP RCERPLQGYS VEYQLLNGGE LHRLNIPNPA QTSVVVEDLL PNHSYVFRVR AQSQEGWGRE AEGVITIESQ VHPQSPLCPL PGSAFTLSTP SAPGPLVFTA LSPDSLQLSW ERPRRPNGDI VGYLVTCEMA QGGGPATAFR VDGDSPESRL TVPGLSENVP YKFKVQARTT EGFGPEREGI ITIES |
-Experimental information
| Beam | Instrument name: PETRA III EMBL P12 / City: Hamburg / 国: Germany  / Type of source: X-ray synchrotron / Wavelength: 0.124 Å / Dist. spec. to detc.: 3.1 mm / Type of source: X-ray synchrotron / Wavelength: 0.124 Å / Dist. spec. to detc.: 3.1 mm | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 2M | ||||||||||||||||||||||||||||||
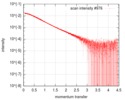
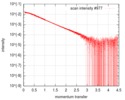
| Scan | Measurement date: Nov 26, 2013 / Cell temperature: 10 °C / Exposure time: 0.05 sec. / Number of frames: 30 / Unit: 1/nm /
| ||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| ||||||||||||||||||||||||||||||
| Result | Comments: 'Synchrotron SAXS data from solutions of a fragment of the cytoplasmic region of the human integrin Beta4 subunit. This fragment includes the final part of a region named the
|
 Movie
Movie Controller
Controller

 SASDDN8
SASDDN8