[English] 日本語
 Yorodumi
Yorodumi- SASDCD3: Nucleolysin TIA-1 isoform p40 (LPQTG containing construct for sor... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 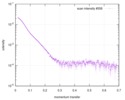 |
|---|---|
 Sample Sample | Nucleolysin TIA-1 isoform p40 (LPQTG containing construct for sortase mediated protein ligation)
|
| Function / homology | Isoform Short of Cytotoxic granule associated RNA binding protein TIA1 Function and homology information Function and homology information |
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
 Citation Citation |  Journal: Angew Chem Int Ed Engl / Year: 2017 Journal: Angew Chem Int Ed Engl / Year: 2017Title: Segmental, Domain-Selective Perdeuteration and Small-Angle Neutron Scattering for Structural Analysis of Multi-Domain Proteins. Authors: Miriam Sonntag / Pravin Kumar Ankush Jagtap / Bernd Simon / Marie-Sousai Appavou / Arie Geerlof / Ralf Stehle / Frank Gabel / Janosch Hennig / Michael Sattler /   Abstract: Multi-domain proteins play critical roles in fine-tuning essential processes in cellular signaling and gene regulation. Typically, multiple globular domains that are connected by flexible linkers ...Multi-domain proteins play critical roles in fine-tuning essential processes in cellular signaling and gene regulation. Typically, multiple globular domains that are connected by flexible linkers undergo dynamic rearrangements upon binding to protein, DNA or RNA ligands. RNA binding proteins (RBPs) represent an important class of multi-domain proteins, which regulate gene expression by recognizing linear or structured RNA sequence motifs. Here, we employ segmental perdeuteration of the three RNA recognition motif (RRM) domains in the RBP TIA-1 using Sortase A mediated protein ligation. We show that domain-selective perdeuteration combined with contrast-matched small-angle neutron scattering (SANS), SAXS and computational modeling provides valuable information to precisely define relative domain arrangements. The approach is generally applicable to study conformational arrangements of individual domains in multi-domain proteins and changes induced by ligand binding. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Models
- Sample
Sample
 Sample Sample | Name: Nucleolysin TIA-1 isoform p40 (LPQTG containing construct for sortase mediated protein ligation) |
|---|---|
| Buffer | Name: 10 mM Potassium Phosphate 50 mM NaCl 10 mM DTT / pH: 6 |
| Entity #661 | Name: TIA-1 C-LPQTG / Type: protein / Description: Nucleolysin TIA-1 isoform p40 / Formula weight: 30.522 / Num. of mol.: 1 / Source: Homo sapiens / References: UniProt: P31483-2 Sequence: MEDEMPKTLY VGNLSRDVTE ALILQLFSQI GPCKNCKMIM DTAGNDPYCF VEFHEHRHAA AALAAMNGRK IMGKEVKVNW ATTPSSQKLP QTGNHFHVFV GDLSPEITTE DIKAAFAPFG RISDARVVKD MATGKSKGYG FVSFFNKWDA ENAIQQMGGQ WLGGRQIRTN ...Sequence: MEDEMPKTLY VGNLSRDVTE ALILQLFSQI GPCKNCKMIM DTAGNDPYCF VEFHEHRHAA AALAAMNGRK IMGKEVKVNW ATTPSSQKLP QTGNHFHVFV GDLSPEITTE DIKAAFAPFG RISDARVVKD MATGKSKGYG FVSFFNKWDA ENAIQQMGGQ WLGGRQIRTN WATRKPPAPK STYESNTKQL SYDEVVNQSS PSNCTVYCGG VTSGLTEQLM RQTFSPFGQI MEIRVFPDKG YSFVRFNSHE SAAHAIVSVN GTTIEGHVVK CYWGK |
-Experimental information
| Beam | Instrument name: Rigaku BioSAXS-1000 / City: Munich / 国: Germany  / Type of source: X-ray in house / Type of source: X-ray in house | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 100K / Pixsize x: 172 mm | ||||||||||||||||||||||||||||||||||||
| Scan | Measurement date: Dec 1, 2015 / Unit: 1/A /
| ||||||||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| ||||||||||||||||||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller


 SASDCD3
SASDCD3






