+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDA57 |
|---|---|
 Sample Sample | Surface Protein G (SasG) EG5 repeat protein G51-G54
|
| Function / homology |  Function and homology information Function and homology information |
| Biological species |  |
 Citation Citation |  Journal: Nat Commun / Year: 2015 Journal: Nat Commun / Year: 2015Title: Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein. Authors: Dominika T Gruszka / Fiona Whelan / Oliver E Farrance / Herman K H Fung / Emanuele Paci / Cy M Jeffries / Dmitri I Svergun / Clair Baldock / Christoph G Baumann / David J Brockwell / ...Authors: Dominika T Gruszka / Fiona Whelan / Oliver E Farrance / Herman K H Fung / Emanuele Paci / Cy M Jeffries / Dmitri I Svergun / Clair Baldock / Christoph G Baumann / David J Brockwell / Jennifer R Potts / Jane Clarke /   Abstract: Bacteria exploit surface proteins to adhere to other bacteria, surfaces and host cells. Such proteins need to project away from the bacterial surface and resist significant mechanical forces. SasG is ...Bacteria exploit surface proteins to adhere to other bacteria, surfaces and host cells. Such proteins need to project away from the bacterial surface and resist significant mechanical forces. SasG is a protein that forms extended fibrils on the surface of Staphylococcus aureus and promotes host adherence and biofilm formation. Here we show that although monomeric and lacking covalent cross-links, SasG maintains a highly extended conformation in solution. This extension is mediated through obligate folding cooperativity of the intrinsically disordered E domains that couple non-adjacent G5 domains thermodynamically, forming interfaces that are more stable than the domains themselves. Thus, counterintuitively, the elongation of the protein appears to be dependent on the inherent instability of its domains. The remarkable mechanical strength of SasG arises from tandemly arrayed 'clamp' motifs within the folded domains. Our findings reveal an elegant minimal solution for the assembly of monomeric mechano-resistant tethers of variable length. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
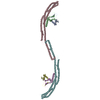
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Models
| Model #239 |  Type: dummy / Software: Gasbor / Radius of dummy atoms: 1.90 A / Chi-square value: 0.9216  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|---|
| Model #240 |  Type: atomic / Software: SASREF / Radius of dummy atoms: 1.90 A / Chi-square value: 0.9409  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
- Sample
Sample
 Sample Sample | Name: Surface Protein G (SasG) EG5 repeat protein G51-G54 / Specimen concentration: 6 mg/ml |
|---|---|
| Buffer | Name: Tris / Concentration: 20.00 mM / pH: 7.5 / Composition: 200 mM NaCl, 1 mM EDTA, 20 mM Tris.Cl |
| Entity #142 | Name: SasG / Type: protein / Description: Surface protein G / Formula weight: 52.802 / Num. of mol.: 1 / Source: Staphylococcus aureus / References: UniProt: Q2G2B2 Sequence: GPAMAPKTIT ELEKKVEEIP FKKERKFNPD LAPGTEKVTR EGQKGEKTIT TPTLKNPLTG VIISKGEPKE EITKDPINEL TEYGPETIAP GHRDEFDPKL PTGEKEEVPG KPGIKNPETG DVVRPPVDSV TKYGPVKGDS IVEKEEIPFE KERKFNPDLA PGTEKVTREG ...Sequence: GPAMAPKTIT ELEKKVEEIP FKKERKFNPD LAPGTEKVTR EGQKGEKTIT TPTLKNPLTG VIISKGEPKE EITKDPINEL TEYGPETIAP GHRDEFDPKL PTGEKEEVPG KPGIKNPETG DVVRPPVDSV TKYGPVKGDS IVEKEEIPFE KERKFNPDLA PGTEKVTREG QKGEKTITTP TLKNPLTGEI ISKGESKEEI TKDPINELTE YGPETITPGH RDEFDPKLPT GEKEEVPGKP GIKNPETGDV VRPPVDSVTK YGPVKGDSIV EKEEIPFEKE RKFNPDLAPG TEKVTREGQK GEKTITTPTL KNPLTGVIIS KGEPKEEITK DPINELTEYG PETITPGHRD EFDPKLPTGE KEEVPGKPGI KNPETGDVVR PPVDSVTKYG PVKGDSIVEK EEIPFKKERK FNPDLAPGTE KVTREGQKGE KTITTPTLKN PLTGEIISKG ESKEEITKDP INELTEYGPE TWSHPQFEK |
-Experimental information
| Beam | Instrument name: PETRA III P12 / City: Hamburg / 国: Germany  / Type of source: X-ray synchrotron / Wavelength: 0.12 Å / Dist. spec. to detc.: 3.1 mm / Type of source: X-ray synchrotron / Wavelength: 0.12 Å / Dist. spec. to detc.: 3.1 mm | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 2M | |||||||||||||||||||||
| Scan |
| |||||||||||||||||||||
| Distance distribution function P(R) |
| |||||||||||||||||||||
| Result | Comments: SAXS was used to determine whether the EG5 repeat forms an extended rod-like structure in solution.
|
 Movie
Movie Controller
Controller


 SASDA57
SASDA57