[English] 日本語
 Yorodumi
Yorodumi- PDB-9hco: Outward-open structure of human serotonin transporter bound to vi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9hco | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Outward-open structure of human serotonin transporter bound to vilazodone | |||||||||||||||||||||||||||
 Components Components |
| |||||||||||||||||||||||||||
 Keywords Keywords | MEMBRANE PROTEIN / Monoamine transporter / LeuT fold / antidepressant binding / Inhibitor complex | |||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of cerebellar granule cell precursor proliferation / regulation of thalamus size / Serotonin clearance from the synaptic cleft / positive regulation of serotonin secretion / serotonergic synapse / negative regulation of synaptic transmission, dopaminergic / cocaine binding / serotonin:sodium:chloride symporter activity / cellular response to cGMP / enteric nervous system development ...negative regulation of cerebellar granule cell precursor proliferation / regulation of thalamus size / Serotonin clearance from the synaptic cleft / positive regulation of serotonin secretion / serotonergic synapse / negative regulation of synaptic transmission, dopaminergic / cocaine binding / serotonin:sodium:chloride symporter activity / cellular response to cGMP / enteric nervous system development / negative regulation of organ growth / sodium ion binding / neurotransmitter transmembrane transporter activity / serotonin uptake / vasoconstriction / monoamine transmembrane transporter activity / monoamine transport / serotonin binding / brain morphogenesis / syntaxin-1 binding / antiporter activity / male mating behavior / negative regulation of neuron differentiation / neurotransmitter transport / nitric-oxide synthase binding / amino acid transport / conditioned place preference / membrane depolarization / monoatomic cation channel activity / behavioral response to cocaine / cellular response to retinoic acid / positive regulation of cell cycle / endomembrane system / response to nutrient / sodium ion transmembrane transport / circadian rhythm / platelet aggregation / response to toxic substance / memory / integrin binding / actin filament binding / response to estradiol / presynaptic membrane / postsynaptic membrane / response to hypoxia / neuron projection / endosome membrane / membrane raft / response to xenobiotic stimulus / focal adhesion / synapse / positive regulation of gene expression / identical protein binding / plasma membrane Similarity search - Function | |||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.78 Å | |||||||||||||||||||||||||||
 Authors Authors | Kalenderoglou, I.E. / Tillmann, P. / Loland, C.J. | |||||||||||||||||||||||||||
| Funding support |  Denmark, European Union, Denmark, European Union,  United States, 7items United States, 7items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structural basis of serotonin reuptake inhibition by a dual mode orthosteric/allosteric antidepressant Authors: Kalenderoglou, I.E. / Nygaard, A. / Vogt, C.D. / Turaev, A. / Pape, T. / Adams, N. / Newman, A.H. / Loland, C.J. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9hco.cif.gz 9hco.cif.gz | 302.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9hco.ent.gz pdb9hco.ent.gz | 243 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  9hco.json.gz 9hco.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  9hco_validation.pdf.gz 9hco_validation.pdf.gz | 1.3 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  9hco_full_validation.pdf.gz 9hco_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  9hco_validation.xml.gz 9hco_validation.xml.gz | 38.7 KB | Display | |
| Data in CIF |  9hco_validation.cif.gz 9hco_validation.cif.gz | 56.4 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/hc/9hco https://data.pdbj.org/pub/pdb/validation_reports/hc/9hco ftp://data.pdbj.org/pub/pdb/validation_reports/hc/9hco ftp://data.pdbj.org/pub/pdb/validation_reports/hc/9hco | HTTPS FTP |
-Related structure data
| Related structure data |  52050MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 76116.453 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: human SERT full length with GGS linker, thromvin site, GGS linker, twinII site, GGS linker and His12-tag Source: (gene. exp.)  Homo sapiens (human) / Gene: SLC6A4, HTT, SERT / Plasmid: pFastBac Homo sapiens (human) / Gene: SLC6A4, HTT, SERT / Plasmid: pFastBacDetails (production host): Baculovirus-derived vector with SLC6A4 gene insert Cell line (production host): Expi293 / Production host:  Homo sapiens (human) / Strain (production host): HEK293 / References: UniProt: P31645 Homo sapiens (human) / Strain (production host): HEK293 / References: UniProt: P31645 |
|---|
-Antibody , 2 types, 2 molecules HL
| #2: Antibody | Mass: 24516.510 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: Murine monoclonal antibody (15B8) specific for the human serotonin transporter (SERT) Source: (gene. exp.)  Details (production host): Baculovirus expression vector used in BacMam system for protein expression in Sf9 cells. Cell (production host): Sf9 / Cell line (production host): Sf9 / Production host:  |
|---|---|
| #3: Antibody | Mass: 23558.977 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: Murine monoclonal antibody (15B8) specific for the human serotonin transporter (SERT) Source: (gene. exp.)  Details (production host): Baculovirus expression vector used in BacMam system for protein expression in Sf9 cells. Cell (production host): Sf9 / Cell line (production host): Sf9 / Production host:  |
-Non-polymers , 4 types, 16 molecules 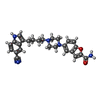






| #4: Chemical | ChemComp-YG7 / | ||||
|---|---|---|---|---|---|
| #5: Chemical | | #6: Chemical | ChemComp-CL / | #7: Water | ChemComp-HOH / | |
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Complex of the Serotonin Transporter with the antidepressant vilazodone Type: COMPLEX / Entity ID: #1-#3 / Source: RECOMBINANT |
|---|---|
| Molecular weight | Experimental value: NO |
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Source (recombinant) | Organism:  Homo sapiens (human) / Strain: HEK293 / Cell: Expi293 / Plasmid: pFastBac Homo sapiens (human) / Strain: HEK293 / Cell: Expi293 / Plasmid: pFastBac |
| Buffer solution | pH: 8 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Specimen support | Grid material: GOLD / Grid mesh size: 300 divisions/in. / Grid type: UltrAuFoil R1.2/1.3 |
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: TFS KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 20000 nm / Nominal defocus min: 6000 nm |
| Specimen holder | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 42 e/Å2 / Detector mode: COUNTING / Film or detector model: FEI FALCON III (4k x 4k) |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 2.78 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 239223 / Symmetry type: POINT |
 Movie
Movie Controller
Controller


 PDBj
PDBj







