[English] 日本語
 Yorodumi
Yorodumi- EMDB-52050: Outward-open structure of human serotonin transporter bound to vi... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Outward-open structure of human serotonin transporter bound to vilazodone | ||||||||||||||||||||||||
 Map data Map data | Sharp EM map | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | Monoamine transporter / LeuT fold / antidepressant binding / Inhibitor complex / MEMBRANE PROTEIN | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of cerebellar granule cell precursor proliferation / regulation of thalamus size / Serotonin clearance from the synaptic cleft / serotonergic synapse / positive regulation of serotonin secretion / negative regulation of synaptic transmission, dopaminergic / cocaine binding / serotonin:sodium:chloride symporter activity / cellular response to cGMP / enteric nervous system development ...negative regulation of cerebellar granule cell precursor proliferation / regulation of thalamus size / Serotonin clearance from the synaptic cleft / serotonergic synapse / positive regulation of serotonin secretion / negative regulation of synaptic transmission, dopaminergic / cocaine binding / serotonin:sodium:chloride symporter activity / cellular response to cGMP / enteric nervous system development / negative regulation of organ growth / sodium ion binding / neurotransmitter transmembrane transporter activity / serotonin uptake / vasoconstriction / monoamine transmembrane transporter activity / monoamine transport / serotonin binding / brain morphogenesis / syntaxin-1 binding / antiporter activity / negative regulation of neuron differentiation / neurotransmitter transport / male mating behavior / nitric-oxide synthase binding / amino acid transport / membrane depolarization / conditioned place preference / monoatomic cation channel activity / behavioral response to cocaine / cellular response to retinoic acid / positive regulation of cell cycle / endomembrane system / response to nutrient / sodium ion transmembrane transport / circadian rhythm / platelet aggregation / integrin binding / response to toxic substance / memory / actin filament binding / response to estradiol / presynaptic membrane / response to hypoxia / postsynaptic membrane / endosome membrane / neuron projection / membrane raft / response to xenobiotic stimulus / focal adhesion / synapse / positive regulation of gene expression / identical protein binding / plasma membrane Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | ||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.78 Å | ||||||||||||||||||||||||
 Authors Authors | Kalenderoglou IE / Tillmann P / Loland CJ | ||||||||||||||||||||||||
| Funding support |  Denmark, European Union, Denmark, European Union,  United States, 7 items United States, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structural basis of serotonin reuptake inhibition by a dual mode orthosteric/allosteric antidepressant Authors: Kalenderoglou IE / Nygaard A / Vogt CD / Turaev A / Pape T / Adams N / Newman AH / Loland CJ | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_52050.map.gz emd_52050.map.gz | 97.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-52050-v30.xml emd-52050-v30.xml emd-52050.xml emd-52050.xml | 24 KB 24 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_52050_fsc.xml emd_52050_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_52050.png emd_52050.png | 102.8 KB | ||
| Filedesc metadata |  emd-52050.cif.gz emd-52050.cif.gz | 7.5 KB | ||
| Others |  emd_52050_additional_1.map.gz emd_52050_additional_1.map.gz emd_52050_half_map_1.map.gz emd_52050_half_map_1.map.gz emd_52050_half_map_2.map.gz emd_52050_half_map_2.map.gz | 50.6 MB 95.4 MB 95.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-52050 http://ftp.pdbj.org/pub/emdb/structures/EMD-52050 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-52050 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-52050 | HTTPS FTP |
-Related structure data
| Related structure data |  9hcoMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_52050.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_52050.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharp EM map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: EM map
| File | emd_52050_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half A
| File | emd_52050_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half B
| File | emd_52050_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of the Serotonin Transporter with the antidepressant vila...
| Entire | Name: Complex of the Serotonin Transporter with the antidepressant vilazodone |
|---|---|
| Components |
|
-Supramolecule #1: Complex of the Serotonin Transporter with the antidepressant vila...
| Supramolecule | Name: Complex of the Serotonin Transporter with the antidepressant vilazodone type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Sodium-dependent serotonin transporter
| Macromolecule | Name: Sodium-dependent serotonin transporter / type: protein_or_peptide / ID: 1 Details: human SERT full length with GGS linker, thromvin site, GGS linker, twinII site, GGS linker and His12-tag Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 76.116453 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: METTPLNSQK QLSACEDGED CQENGVLQKV VPTPGDKVES GQISNGYSAV PSPGAGDDTR HSIPATTTTL VAELHQGERE TWGKKVDFL LSVIGYAVDL GNVWRFPYIC YQNGGGAFLL PYTIMAIFGG IPLFYMELAL GQYHRNGCIS IWRKICPIFK G IGYAICII ...String: METTPLNSQK QLSACEDGED CQENGVLQKV VPTPGDKVES GQISNGYSAV PSPGAGDDTR HSIPATTTTL VAELHQGERE TWGKKVDFL LSVIGYAVDL GNVWRFPYIC YQNGGGAFLL PYTIMAIFGG IPLFYMELAL GQYHRNGCIS IWRKICPIFK G IGYAICII AFYIASYYNT IMAWALYYLI SSFTDQLPWT SCKNSWNTGN CTNYFSEDNI TWTLHSTSPA EEFYTRHVLQ IH RSKGLQD LGGISWQLAL CIMLIFTVIY FSIWKGVKTS GKVVWVTATF PYIILSVLLV RGATLPGAWR GVLFYLKPNW QKL LETGVW IDAAAQIFFS LGPGFGVLLA FASYNKFNNN CYQDALVTSV VNCMTSFVSG FVIFTVLGYM AEMRNEDVSE VAKD AGPSL LFITYAEAIA NMPASTFFAI IFFLMLITLG LDSTFAGLEG VITAVLDEFP HVWAKRRERF VLAVVITCFF GSLVT LTFG GAYVVKLLEE YATGPAVLTV ALIEAVAVSW FYGITQFCRD VKEMLGFSPG WFWRICWVAI SPLFLLFIIC SFLMSP PQL RLFQYNYPYW SIILGYCIGT SSFICIPTYI AYRLIITPGT FKERIIKSIT PETPTEIPCG DIRLNAVGGS LVPRGSG GS WSHPQFEKGG GSGGGSGGSA WSHPQFEKGG SHHHHHHHHH HHH UniProtKB: Sodium-dependent serotonin transporter |
-Macromolecule #2: 15B8 Fab heavy chain
| Macromolecule | Name: 15B8 Fab heavy chain / type: protein_or_peptide / ID: 2 / Details: Antibody heavy chain Fab with His6 tag / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.51651 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQQSGPE LVKLGASVRI SCKASGYRFS YSWMNWVKQR PGKGLEWIGR IYPGDGDTKY SGKFKGKATL TADKSSSTVY MQLSSLTSE DSAVYFCARS AYGSEGFAMD YWGQGTSVTV SSAKTTAPSV FPLAPVCGDT TGSSVTLGCL VKGYFPEPVT L TWNSGSLS ...String: QVQLQQSGPE LVKLGASVRI SCKASGYRFS YSWMNWVKQR PGKGLEWIGR IYPGDGDTKY SGKFKGKATL TADKSSSTVY MQLSSLTSE DSAVYFCARS AYGSEGFAMD YWGQGTSVTV SSAKTTAPSV FPLAPVCGDT TGSSVTLGCL VKGYFPEPVT L TWNSGSLS SGVHTFPAVL QSGLYTLSSS VTVTSSTWPS QSITCNVAHP ASSTKVDKKL VPRGSHHHHH H |
-Macromolecule #3: 15B8 Fab light chain
| Macromolecule | Name: 15B8 Fab light chain / type: protein_or_peptide / ID: 3 / Details: Antibody ligh chain Fab / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.558977 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DIVLTQSPAS LAVSLGQRAT ISCRASESVD NYGISFLNWF QQKPGQPPKL LIYAASNQGS GVPARFSGSG SGTYFSLNIH PMEEDDTAV YFCQQTKGVS WTFGGGTKVE IKRADAAPTV SVFPPSSEQL TSGGASVVCF LNNFYPRDIN VKWKIDGSER Q NGVLNSWT ...String: DIVLTQSPAS LAVSLGQRAT ISCRASESVD NYGISFLNWF QQKPGQPPKL LIYAASNQGS GVPARFSGSG SGTYFSLNIH PMEEDDTAV YFCQQTKGVS WTFGGGTKVE IKRADAAPTV SVFPPSSEQL TSGGASVVCF LNNFYPRDIN VKWKIDGSER Q NGVLNSWT DQDSKDSTYS MSSTLTLTKD EYERHNSYTC EATHKTSTSP IVKSFNRG |
-Macromolecule #4: 5-{4-[4-(5-cyano-1H-indol-3-yl)butyl]piperazin-1-yl}-1-benzofuran...
| Macromolecule | Name: 5-{4-[4-(5-cyano-1H-indol-3-yl)butyl]piperazin-1-yl}-1-benzofuran-2-carboxamide type: ligand / ID: 4 / Number of copies: 1 / Formula: YG7 |
|---|---|
| Molecular weight | Theoretical: 441.525 Da |
| Chemical component information | 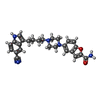 ChemComp-YG7: |
-Macromolecule #5: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 5 / Number of copies: 2 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #6: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 12 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 20.0 µm / Nominal defocus min: 6.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)














































