+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8urj | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
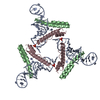
| Title | Cryo-EM structure of the HIV-1 nuclear export complex | |||||||||||||||||||||||||||
 Components Components |
| |||||||||||||||||||||||||||
 Keywords Keywords | RNA BINDING PROTEIN/RNA / HIV Karyopherin Rev Crm1 / RNA BINDING PROTEIN-RNA complex | |||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcellular response to triglyceride / cellular response to salt / HuR (ELAVL1) binds and stabilizes mRNA / annulate lamellae / regulation of proteasomal ubiquitin-dependent protein catabolic process / pre-miRNA export from nucleus / RNA nuclear export complex / snRNA import into nucleus / manchette / nuclear export signal receptor activity ...cellular response to triglyceride / cellular response to salt / HuR (ELAVL1) binds and stabilizes mRNA / annulate lamellae / regulation of proteasomal ubiquitin-dependent protein catabolic process / pre-miRNA export from nucleus / RNA nuclear export complex / snRNA import into nucleus / manchette / nuclear export signal receptor activity / regulation of centrosome duplication / cellular response to mineralocorticoid stimulus / Regulation of cholesterol biosynthesis by SREBP (SREBF) / importin-alpha family protein binding / regulation of protein export from nucleus / Rev-mediated nuclear export of HIV RNA / Nuclear import of Rev protein / protein localization to nucleolus / NEP/NS2 Interacts with the Cellular Export Machinery / tRNA processing in the nucleus / GTP metabolic process / Postmitotic nuclear pore complex (NPC) reformation / nucleocytoplasmic transport / MicroRNA (miRNA) biogenesis / DNA metabolic process / dynein intermediate chain binding / Maturation of hRSV A proteins / mitotic sister chromatid segregation / viral process / spermatid development / ribosomal large subunit export from nucleus / Estrogen-dependent nuclear events downstream of ESR-membrane signaling / protein localization to nucleus / nuclear pore / sperm flagellum / positive regulation of protein binding / mRNA export from nucleus / Cajal body / Cyclin A/B1/B2 associated events during G2/M transition / ribosomal subunit export from nucleus / NPAS4 regulates expression of target genes / ribosomal small subunit export from nucleus / Amplification of signal from unattached kinetochores via a MAD2 inhibitory signal / centriole / Transcriptional and post-translational regulation of MITF-M expression and activity / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / protein export from nucleus / Resolution of Sister Chromatid Cohesion / Downregulation of TGF-beta receptor signaling / mitotic spindle organization / male germ cell nucleus / hippocampus development / Deactivation of the beta-catenin transactivating complex / Heme signaling / Transcriptional regulation by small RNAs / RHO GTPases Activate Formins / recycling endosome / MAPK6/MAPK4 signaling / kinetochore / positive regulation of protein import into nucleus / small GTPase binding / protein import into nucleus / Separation of Sister Chromatids / GDP binding / melanosome / nuclear envelope / mitotic cell cycle / ribosome biogenesis / G protein activity / actin cytoskeleton organization / midbody / nuclear membrane / DNA-binding transcription factor binding / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / cadherin binding / response to xenobiotic stimulus / protein heterodimerization activity / ribonucleoprotein complex / protein domain specific binding / cell division / intracellular membrane-bounded organelle / GTPase activity / chromatin binding / chromatin / GTP binding / protein-containing complex binding / nucleolus / magnesium ion binding / negative regulation of transcription by RNA polymerase II / protein-containing complex / RNA binding / extracellular exosome / nucleoplasm / nucleus / membrane / cytosol / cytoplasm Similarity search - Function | |||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human)  Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 4.25 Å | |||||||||||||||||||||||||||
 Authors Authors | Smith, A.M. / Cheng, Y. / Frankel, A.D. | |||||||||||||||||||||||||||
| Funding support |  United States, 5items United States, 5items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure of the HIV-1 RNA Nuclear Export Complex Reveals Crm1 Versatility Authors: Smith, A.M. / Cheng, Y. / Frankel, A.D. / Li, Y. / Velarde, A. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8urj.cif.gz 8urj.cif.gz | 856.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8urj.ent.gz pdb8urj.ent.gz | 700.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8urj.json.gz 8urj.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ur/8urj https://data.pdbj.org/pub/pdb/validation_reports/ur/8urj ftp://data.pdbj.org/pub/pdb/validation_reports/ur/8urj ftp://data.pdbj.org/pub/pdb/validation_reports/ur/8urj | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  42494MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 122267.984 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: XPO1 / Production host: Homo sapiens (human) / Gene: XPO1 / Production host:  Homo sapiens (human) / Strain (production host): HEK293-F / References: UniProt: O14980 Homo sapiens (human) / Strain (production host): HEK293-F / References: UniProt: O14980#2: Protein | | Mass: 10687.159 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: Native NES was exchanged with NS2Val NES Native NES: LPPLERLTL Ns2Val NES: TVDEMTKKFGTLTI Source: (gene. exp.)   Human immunodeficiency virus 1 / Strain: HXB3 / Gene: Rev / Production host: Human immunodeficiency virus 1 / Strain: HXB3 / Gene: Rev / Production host:  #3: Protein | Mass: 20722.076 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Details: Q69L 1-180 / Source: (gene. exp.)  Homo sapiens (human) / Gene: RAN, ARA24, OK/SW-cl.81 / Production host: Homo sapiens (human) / Gene: RAN, ARA24, OK/SW-cl.81 / Production host:  References: UniProt: P62826, Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement #4: Protein | | Mass: 10644.108 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: Native NES was exchanged with NS2Val NES Native NES: LPPLERLTL Ns2Val NES: TVDEMTKKFGTLTI Source: (gene. exp.)   Human immunodeficiency virus 1 / Strain: HXB3 / Production host: Human immunodeficiency virus 1 / Strain: HXB3 / Production host:  #5: RNA chain | | Mass: 114996.102 Da / Num. of mol.: 1 / Mutation: G262A G269A / Source method: obtained synthetically / Source: (synth.)   Human immunodeficiency virus 1 / References: GenBank: 328658 Human immunodeficiency virus 1 / References: GenBank: 328658Has ligand of interest | Y | Has protein modification | N | |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component |
| |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight |
| |||||||||||||||||||||||||||||||||||
| Source (natural) |
| |||||||||||||||||||||||||||||||||||
| Source (recombinant) |
| |||||||||||||||||||||||||||||||||||
| Buffer solution | pH: 7.5 | |||||||||||||||||||||||||||||||||||
| Buffer component |
| |||||||||||||||||||||||||||||||||||
| Specimen | Conc.: 0.7 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | |||||||||||||||||||||||||||||||||||
| Specimen support | Grid material: GOLD / Grid mesh size: 300 divisions/in. / Grid type: Quantifoil R1.2/1.3 | |||||||||||||||||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 298.15 K Details: Washed grid 2x outside of vitrobot; with the second drop of buffer I loaded the tweezers; moved the grid into the chamber and then I blotted. |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Microscopy | Model: FEI TITAN KRIOS | |||||||||||||||||||||
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM | |||||||||||||||||||||
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 105000 X / Calibrated magnification: 59880 X / Nominal defocus max: 2000 nm / Nominal defocus min: 800 nm / Cs: 2.7 mm / C2 aperture diameter: 100 µm / Alignment procedure: COMA FREE | |||||||||||||||||||||
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER | |||||||||||||||||||||
| Image recording |
| |||||||||||||||||||||
| EM imaging optics | Energyfilter name: GIF Bioquantum / Energyfilter slit width: 20 eV |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 11176625 | ||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 4.25 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 260885 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | B value: 325 / Protocol: RIGID BODY FIT | ||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | 3D fitting-ID: 1 / Source name: PDB / Type: experimental model
| ||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj