[English] 日本語
 Yorodumi
Yorodumi- PDB-8gz2: Cryo-EM structure of human NaV1.6/beta1/beta2-4,9-anhydro-tetrodotoxin -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8gz2 | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human NaV1.6/beta1/beta2-4,9-anhydro-tetrodotoxin | |||||||||||||||||||||||||||||||||
 Components Components |
| |||||||||||||||||||||||||||||||||
 Keywords Keywords | MEMBRANE PROTEIN / ion channal | |||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to pyrethroid / corticospinal neuron axon guidance / positive regulation of voltage-gated sodium channel activity / voltage-gated sodium channel activity involved in cardiac muscle cell action potential / regulation of atrial cardiac muscle cell membrane depolarization / voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization / membrane depolarization during Purkinje myocyte cell action potential / sodium ion binding / cardiac conduction / membrane depolarization during cardiac muscle cell action potential ...response to pyrethroid / corticospinal neuron axon guidance / positive regulation of voltage-gated sodium channel activity / voltage-gated sodium channel activity involved in cardiac muscle cell action potential / regulation of atrial cardiac muscle cell membrane depolarization / voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization / membrane depolarization during Purkinje myocyte cell action potential / sodium ion binding / cardiac conduction / membrane depolarization during cardiac muscle cell action potential / membrane depolarization during action potential / positive regulation of sodium ion transport / regulation of sodium ion transmembrane transport / peripheral nervous system development / axon initial segment / regulation of ventricular cardiac muscle cell membrane repolarization / cardiac muscle cell action potential involved in contraction / node of Ranvier / voltage-gated sodium channel complex / sodium channel inhibitor activity / locomotion / neuronal action potential propagation / anchoring junction / podosome / Interaction between L1 and Ankyrins / voltage-gated sodium channel activity / sodium ion transport / Phase 0 - rapid depolarisation / regulation of heart rate by cardiac conduction / action potential / intercalated disc / sodium channel regulator activity / membrane depolarization / cardiac muscle contraction / myelination / T-tubule / axon guidance / sodium ion transmembrane transport / positive regulation of neuron projection development / Z disc / Sensory perception of sweet, bitter, and umami (glutamate) taste / cell junction / nervous system development / response to heat / cytoplasmic vesicle / gene expression / chemical synaptic transmission / perikaryon / transmembrane transporter binding / cell adhesion / axon / synapse / extracellular region / membrane / plasma membrane Similarity search - Function | |||||||||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||||||||||||||||||||||||||
 Authors Authors | Li, Y. / Jiang, D. | |||||||||||||||||||||||||||||||||
| Funding support |  China, 1items China, 1items
| |||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structure of human Na1.6 channel reveals Na selectivity and pore blockade by 4,9-anhydro-tetrodotoxin. Authors: Yue Li / Tian Yuan / Bo Huang / Feng Zhou / Chao Peng / Xiaojing Li / Yunlong Qiu / Bei Yang / Yan Zhao / Zhuo Huang / Daohua Jiang /  Abstract: The sodium channel Na1.6 is widely expressed in neurons of the central and peripheral nervous systems, which plays a critical role in regulating neuronal excitability. Dysfunction of Na1.6 has been ...The sodium channel Na1.6 is widely expressed in neurons of the central and peripheral nervous systems, which plays a critical role in regulating neuronal excitability. Dysfunction of Na1.6 has been linked to epileptic encephalopathy, intellectual disability and movement disorders. Here we present cryo-EM structures of human Na1.6/β1/β2 alone and complexed with a guanidinium neurotoxin 4,9-anhydro-tetrodotoxin (4,9-ah-TTX), revealing molecular mechanism of Na1.6 inhibition by the blocker. The apo-form structure reveals two potential Na binding sites within the selectivity filter, suggesting a possible mechanism for Na selectivity and conductance. In the 4,9-ah-TTX bound structure, 4,9-ah-TTX binds to a pocket similar to the tetrodotoxin (TTX) binding site, which occupies the Na binding sites and completely blocks the channel. Molecular dynamics simulation results show that subtle conformational differences in the selectivity filter affect the affinity of TTX analogues. Taken together, our results provide important insights into Na1.6 structure, ion conductance, and inhibition. | |||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8gz2.cif.gz 8gz2.cif.gz | 305.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8gz2.ent.gz pdb8gz2.ent.gz | 230.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8gz2.json.gz 8gz2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/gz/8gz2 https://data.pdbj.org/pub/pdb/validation_reports/gz/8gz2 ftp://data.pdbj.org/pub/pdb/validation_reports/gz/8gz2 ftp://data.pdbj.org/pub/pdb/validation_reports/gz/8gz2 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  34388MC  8gz1C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Sodium channel subunit beta- ... , 2 types, 2 molecules DC
| #1: Protein | Mass: 24732.115 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: SCN1B / Cell line (production host): HEK293 / Production host: Homo sapiens (human) / Gene: SCN1B / Cell line (production host): HEK293 / Production host:  Homo sapiens (human) / References: UniProt: Q07699 Homo sapiens (human) / References: UniProt: Q07699 |
|---|---|
| #3: Protein | Mass: 24355.859 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: SCN2B, UNQ326/PRO386 / Production host: Homo sapiens (human) / Gene: SCN2B, UNQ326/PRO386 / Production host:  Homo sapiens (human) / References: UniProt: O60939 Homo sapiens (human) / References: UniProt: O60939 |
-Protein / Non-polymers , 2 types, 2 molecules B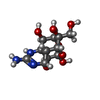

| #2: Protein | Mass: 225520.609 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: SCN8A, MED / Production host: Homo sapiens (human) / Gene: SCN8A, MED / Production host:  Homo sapiens (human) / References: UniProt: Q9UQD0 Homo sapiens (human) / References: UniProt: Q9UQD0 |
|---|---|
| #6: Chemical | ChemComp-WMK / ( |
-Sugars , 2 types, 7 molecules 
| #4: Polysaccharide | beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta- ...beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose Source method: isolated from a genetically manipulated source #5: Sugar | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Structure of human voltage-gated sodium channel Nav1.6 in complex with auxiliary beta subunits Type: COMPLEX / Entity ID: #1-#3 / Source: RECOMBINANT |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Source (recombinant) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Buffer solution | pH: 7.5 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2200 nm / Nominal defocus min: 1200 nm / Cs: 2.7 mm |
| Image recording | Electron dose: 9.2 e/Å2 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.18.2_3874: / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software | Name: PHENIX / Category: model refinement | ||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING ONLY | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.3 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 76448 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj






