[English] 日本語
 Yorodumi
Yorodumi- EMDB-34388: Cryo-EM structure of human NaV1.6/beta1/beta2-4,9-anhydro-tetrodotoxin -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human NaV1.6/beta1/beta2-4,9-anhydro-tetrodotoxin | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ion channal / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to pyrethroid / corticospinal neuron axon guidance / positive regulation of voltage-gated sodium channel activity / voltage-gated sodium channel activity involved in cardiac muscle cell action potential / regulation of atrial cardiac muscle cell membrane depolarization / voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization / membrane depolarization during Purkinje myocyte cell action potential / sodium ion binding / cardiac conduction / membrane depolarization during cardiac muscle cell action potential ...response to pyrethroid / corticospinal neuron axon guidance / positive regulation of voltage-gated sodium channel activity / voltage-gated sodium channel activity involved in cardiac muscle cell action potential / regulation of atrial cardiac muscle cell membrane depolarization / voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization / membrane depolarization during Purkinje myocyte cell action potential / sodium ion binding / cardiac conduction / membrane depolarization during cardiac muscle cell action potential / membrane depolarization during action potential / positive regulation of sodium ion transport / regulation of sodium ion transmembrane transport / peripheral nervous system development / axon initial segment / regulation of ventricular cardiac muscle cell membrane repolarization / cardiac muscle cell action potential involved in contraction / node of Ranvier / voltage-gated sodium channel complex / sodium channel inhibitor activity / locomotion / neuronal action potential propagation / anchoring junction / podosome / Interaction between L1 and Ankyrins / voltage-gated sodium channel activity / sodium ion transport / Phase 0 - rapid depolarisation / regulation of heart rate by cardiac conduction / action potential / intercalated disc / sodium channel regulator activity / membrane depolarization / cardiac muscle contraction / myelination / T-tubule / axon guidance / sodium ion transmembrane transport / positive regulation of neuron projection development / Z disc / Sensory perception of sweet, bitter, and umami (glutamate) taste / cell junction / nervous system development / response to heat / cytoplasmic vesicle / gene expression / chemical synaptic transmission / perikaryon / transmembrane transporter binding / cell adhesion / axon / synapse / extracellular region / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Li Y / Jiang D | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structure of human Na1.6 channel reveals Na selectivity and pore blockade by 4,9-anhydro-tetrodotoxin. Authors: Yue Li / Tian Yuan / Bo Huang / Feng Zhou / Chao Peng / Xiaojing Li / Yunlong Qiu / Bei Yang / Yan Zhao / Zhuo Huang / Daohua Jiang /  Abstract: The sodium channel Na1.6 is widely expressed in neurons of the central and peripheral nervous systems, which plays a critical role in regulating neuronal excitability. Dysfunction of Na1.6 has been ...The sodium channel Na1.6 is widely expressed in neurons of the central and peripheral nervous systems, which plays a critical role in regulating neuronal excitability. Dysfunction of Na1.6 has been linked to epileptic encephalopathy, intellectual disability and movement disorders. Here we present cryo-EM structures of human Na1.6/β1/β2 alone and complexed with a guanidinium neurotoxin 4,9-anhydro-tetrodotoxin (4,9-ah-TTX), revealing molecular mechanism of Na1.6 inhibition by the blocker. The apo-form structure reveals two potential Na binding sites within the selectivity filter, suggesting a possible mechanism for Na selectivity and conductance. In the 4,9-ah-TTX bound structure, 4,9-ah-TTX binds to a pocket similar to the tetrodotoxin (TTX) binding site, which occupies the Na binding sites and completely blocks the channel. Molecular dynamics simulation results show that subtle conformational differences in the selectivity filter affect the affinity of TTX analogues. Taken together, our results provide important insights into Na1.6 structure, ion conductance, and inhibition. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34388.map.gz emd_34388.map.gz | 59.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34388-v30.xml emd-34388-v30.xml emd-34388.xml emd-34388.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34388.png emd_34388.png | 127.3 KB | ||
| Filedesc metadata |  emd-34388.cif.gz emd-34388.cif.gz | 7.6 KB | ||
| Others |  emd_34388_half_map_1.map.gz emd_34388_half_map_1.map.gz emd_34388_half_map_2.map.gz emd_34388_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34388 http://ftp.pdbj.org/pub/emdb/structures/EMD-34388 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34388 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34388 | HTTPS FTP |
-Related structure data
| Related structure data |  8gz2MC  8gz1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34388.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34388.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
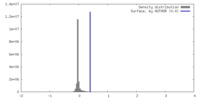
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34388_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
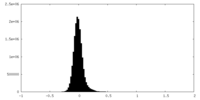
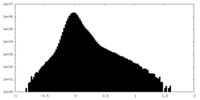
| Density Histograms |
-Half map: #1
| File | emd_34388_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
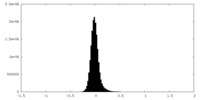
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of human voltage-gated sodium channel Nav1.6 in complex...
| Entire | Name: Structure of human voltage-gated sodium channel Nav1.6 in complex with auxiliary beta subunits |
|---|---|
| Components |
|
-Supramolecule #1: Structure of human voltage-gated sodium channel Nav1.6 in complex...
| Supramolecule | Name: Structure of human voltage-gated sodium channel Nav1.6 in complex with auxiliary beta subunits type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Sodium channel subunit beta-1
| Macromolecule | Name: Sodium channel subunit beta-1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.732115 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGRLLALVVG AALVSSACGG CVEVDSETEA VYGMTFKILC ISCKRRSETN AETFTEWTFR QKGTEEFVKI LRYENEVLQL EEDERFEGR VVWNGSRGTK DLQDLSIFIT NVTYNHSGDY ECHVYRLLFF ENYEHNTSVV KKIHIEVVDK ANRDMASIVS E IMMYVLIV ...String: MGRLLALVVG AALVSSACGG CVEVDSETEA VYGMTFKILC ISCKRRSETN AETFTEWTFR QKGTEEFVKI LRYENEVLQL EEDERFEGR VVWNGSRGTK DLQDLSIFIT NVTYNHSGDY ECHVYRLLFF ENYEHNTSVV KKIHIEVVDK ANRDMASIVS E IMMYVLIV VLTIWLVAEM IYCYKKIAAA TETAAQENAS EYLAITSESK ENCTGVQVAE UniProtKB: Sodium channel regulatory subunit beta-1 |
-Macromolecule #2: Sodium channel protein type 8 subunit alpha
| Macromolecule | Name: Sodium channel protein type 8 subunit alpha / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 225.520609 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAARLLAPPG PDSFKPFTPE SLANIERRIA ESKLKKPPKA DGSHREDDED SKPKPNSDLE AGKSLPFIYG DIPQGLVAVP LEDFDPYYL TQKTFVVLNR GKTLFRFSAT PALYILSPFN LIRRIAIKIL IHSVFSMIIM CTILTNCVFM TFSNPPDWSK N VEYTFTGI ...String: MAARLLAPPG PDSFKPFTPE SLANIERRIA ESKLKKPPKA DGSHREDDED SKPKPNSDLE AGKSLPFIYG DIPQGLVAVP LEDFDPYYL TQKTFVVLNR GKTLFRFSAT PALYILSPFN LIRRIAIKIL IHSVFSMIIM CTILTNCVFM TFSNPPDWSK N VEYTFTGI YTFESLVKII ARGFCIDGFT FLRDPWNWLD FSVIMMAYIT EFVNLGNVSA LRTFRVLRAL KTISVIPGLK TI VGALIQS VKKLSDVMIL TVFCLSVFAL IGLQLFMGNL RNKCVVWPIN FNESYLENGT KGFDWEEYIN NKTNFYTVPG MLE PLLCGN SSDAGQCPEG YQCMKAGRNP NYGYTSFDTF SWAFLALFRL MTQDYWENLY QLTLRAAGKT YMIFFVLVIF VGSF YLVNL ILAVVAMAYE EQNQATLEEA EQKEAEFKAM LEQLKKQQEE AQAAAMATSA GTVSEDAIEE EGEEGGGSPR SSSEI SKLS SKSAKERRNR RKKRKQKELS EGEEKGDPEK VFKSESEDGM RRKAFRLPDN RIGRKFSIMN QSLLSIPGSP FLSRHN SKS SIFSFRGPGR FRDPGSENEF ADDEHSTVEE SEGRRDSLFI PIRARERRSS YSGYSGYSQG SRSSRIFPSL RRSVKRN ST VDCNGVVSLI GGPGSHIGGR LLPEATTEVE IKKKGPGSLL VSMDQLASYG RKDRINSIMS VVTNTLVEEL EESQRKCP P CWYKFANTFL IWECHPYWIK LKEIVNLIVM DPFVDLAITI CIVLNTLFMA MEHHPMTPQF EHVLAVGNLV FTGIFTAEM FLKLIAMDPY YYFQEGWNIF DGFIVSLSLM ELSLADVEGL SVLRSFRLLR VFKLAKSWPT LNMLIKIIGN SVGALGNLTL VLAIIVFIF AVVGMQLFGK SYKECVCKIN QDCELPRWHM HDFFHSFLIV FRVLCGEWIE TMWDCMEVAG QAMCLIVFMM V MVIGNLVV LNLFLALLLS SFSADNLAAT DDDGEMNNLQ ISVIRIKKGV AWTKLKVHAF MQAHFKQREA DEVKPLDELY EK KANCIAN HTGADIHRNG DFQKNGNGTT SGIGSSVEKY IIDEDHMSFI NNPNLTVRVP IAVGESDFEN LNTEDVSSES DPE GSKDKL DDTSSSEGST IDIKPEVEEV PVEQPEEYLD PDACFTEGCV QRFKCCQVNI EEGLGKSWWI LRKTCFLIVE HNWF ETFII FMILLSSGAL AFEDIYIEQR KTIRTILEYA DKVFTYIFIL EMLLKWTAYG FVKFFTNAWC WLDFLIVAVS LVSLI ANAL GYSELGAIKS LRTLRALRPL RALSRFEGMR VVVNALVGAI PSIMNVLLVC LIFWLIFSIM GVNLFAGKYH YCFNET SEI RFEIEDVNNK TECEKLMEGN NTEIRWKNVK INFDNVGAGY LALLQVATFK GWMDIMYAAV DSRKPDEQPK YEDNIYM YI YFVIFIIFGS FFTLNLFIGV IIDNFNQQKK KFGGQDIFMT EEQKKYYNAM KKLGSKKPQK PIPRPLNKIQ GIVFDFVT Q QAFDIVIMML ICLNMVTMMV ETDTQSKQME NILYWINLVF VIFFTCECVL KMFALRHYYF TIGWNIFDFV VVILSIVGM FLADIIEKYF VSPTLFRVIR LARIGRILRL IKGAKGIRTL LFALMMSLPA LFNIGLLLFL VMFIFSIFGM SNFAYVKHEA GIDDMFNFE TFGNSMICLF QITTSAGWDG LLLPILNRPP DCSLDKEHPG SGFKGDCGNP SVGIFFFVSY IIISFLIVVN M YIAIILEN FSVATEESAD PLSEDDFETF YEIWEKFDPD ATQFIEYCKL ADFADALEHP LRVPKPNTIE LIAMDLPMVS GD RIHCLDI LFAFTKRVLG DSGELDILRQ QMEERFVASN PSKVSYEPIT TTLRRKQEEV SAVVLQRAYR GHLARRGFIC KKT TSNKLE NGGTHREKKE STPSTASLPS YDSVTKPEKE KQQRAEEGRR ERAKRQKEVR ESKC UniProtKB: Sodium channel protein type 8 subunit alpha |
-Macromolecule #3: Sodium channel subunit beta-2
| Macromolecule | Name: Sodium channel subunit beta-2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.355859 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MHRDAWLPRP AFSLTGLSLF FSLVPPGRSM EVTVPATLNV LNGSDARLPC TFNSCYTVNH KQFSLNWTYQ ECNNCSEEMF LQFRMKIIN LKLERFQDRV EFSGNPSKYD VSVMLRNVQP EDEGIYNCYI MNPPDRHRGH GKIHLQVLME EPPERDSTVA V IVGASVGG ...String: MHRDAWLPRP AFSLTGLSLF FSLVPPGRSM EVTVPATLNV LNGSDARLPC TFNSCYTVNH KQFSLNWTYQ ECNNCSEEMF LQFRMKIIN LKLERFQDRV EFSGNPSKYD VSVMLRNVQP EDEGIYNCYI MNPPDRHRGH GKIHLQVLME EPPERDSTVA V IVGASVGG FLAVVILVLM VVKCVRRKKE QKLSTDDLKT EEEGKTDGEG NPDDGAK UniProtKB: Sodium channel regulatory subunit beta-2 |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #6: (1R,2S,3S,4R,5R,9S,11S,12S,14R)-7-amino-2,4,12-trihydroxy-2-(hydr...
| Macromolecule | Name: (1R,2S,3S,4R,5R,9S,11S,12S,14R)-7-amino-2,4,12-trihydroxy-2-(hydroxymethyl)-10,13,15-trioxa-6,8-diazapentacyclo[7.4.1.1~3,12~.0~5,11~.0~5,14~]pentadec-7-en-8-ium (non-preferred name) type: ligand / ID: 6 / Number of copies: 1 / Formula: WMK |
|---|---|
| Molecular weight | Theoretical: 301.253 Da |
| Chemical component information | 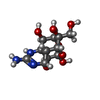 ChemComp-WMK: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 9.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)