+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6el1 | ||||||
|---|---|---|---|---|---|---|---|
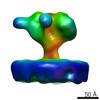
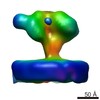
| Title | YaxAB pore complex | ||||||
 Components Components |
| ||||||
 Keywords Keywords | MEMBRANE PROTEIN / Pore-forming toxin / Pathogens / Two-component toxin | ||||||
| Function / homology | : / membrane => GO:0016020 / membrane / Uncharacterized protein / Membrane protein / Alpha-xenorhabdolysin family binary toxin subunit B Function and homology information Function and homology information | ||||||
| Biological species |  Yersinia enterocolitica (bacteria) Yersinia enterocolitica (bacteria) | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 6.1 Å | ||||||
 Authors Authors | Braeuning, B. / Bertosin, E. / Dietz, H. / Groll, M. | ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: Structure and mechanism of the two-component α-helical pore-forming toxin YaxAB. Authors: Bastian Bräuning / Eva Bertosin / Florian Praetorius / Christian Ihling / Alexandra Schatt / Agnes Adler / Klaus Richter / Andrea Sinz / Hendrik Dietz / Michael Groll /  Abstract: Pore-forming toxins (PFT) are virulence factors that transform from soluble to membrane-bound states. The Yersinia YaxAB system represents a family of binary α-PFTs with orthologues in human, ...Pore-forming toxins (PFT) are virulence factors that transform from soluble to membrane-bound states. The Yersinia YaxAB system represents a family of binary α-PFTs with orthologues in human, insect, and plant pathogens, with unknown structures. YaxAB was shown to be cytotoxic and likely involved in pathogenesis, though the molecular basis for its two-component lytic mechanism remains elusive. Here, we present crystal structures of YaxA and YaxB, together with a cryo-electron microscopy map of the YaxAB complex. Our structures reveal a pore predominantly composed of decamers of YaxA-YaxB heterodimers. Both subunits bear membrane-active moieties, but only YaxA is capable of binding to membranes by itself. YaxB can subsequently be recruited to membrane-associated YaxA and induced to present its lytic transmembrane helices. Pore formation can progress by further oligomerization of YaxA-YaxB dimers. Our results allow for a comparison between pore assemblies belonging to the wider ClyA-like family of α-PFTs, highlighting diverse pore architectures. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6el1.cif.gz 6el1.cif.gz | 1.1 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6el1.ent.gz pdb6el1.ent.gz | 977.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6el1.json.gz 6el1.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/el/6el1 https://data.pdbj.org/pub/pdb/validation_reports/el/6el1 ftp://data.pdbj.org/pub/pdb/validation_reports/el/6el1 ftp://data.pdbj.org/pub/pdb/validation_reports/el/6el1 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3885MC  6ek4C  6ek7C  6ek8C C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 45877.156 Da / Num. of mol.: 10 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Yersinia enterocolitica (bacteria) / Gene: ERS137951_00706 / Production host: Yersinia enterocolitica (bacteria) / Gene: ERS137951_00706 / Production host:  #2: Protein | Mass: 39248.836 Da / Num. of mol.: 10 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Yersinia enterocolitica (bacteria) / Gene: YE1985 / Production host: Yersinia enterocolitica (bacteria) / Gene: YE1985 / Production host:  |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: YaxAB complex (10x YaxA + 10x YaxB) / Type: COMPLEX Details: Purified with Cymal-6 detergent and reconstituted in amphipol prior to Cryo-EM. Entity ID: all / Source: RECOMBINANT | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Value: 0.85 MDa / Experimental value: YES | |||||||||||||||
| Source (natural) | Organism:  Yersinia enterocolitica (bacteria) Yersinia enterocolitica (bacteria) | |||||||||||||||
| Source (recombinant) | Organism:  | |||||||||||||||
| Buffer solution | pH: 7 | |||||||||||||||
| Buffer component |
| |||||||||||||||
| Specimen | Conc.: 2 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES Details: Sample exchanged to amphibole A8-35 and run in final round of gel filtration (buffer: 20 mM HEPES pH 7.0, 100 mM NaCl) prior to Cryo-EM. | |||||||||||||||
| Specimen support | Grid material: COPPER / Grid mesh size: 400 divisions/in. / Grid type: C-flat-2/1 | |||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 20 K Details: 3 mM F-FOS Choline 8 just before vitrification blot for 3 to 4 s blot distance -2 to -1 mm |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD |
| Image recording | Electron dose: 60 e/Å2 / Detector mode: INTEGRATING / Film or detector model: FEI FALCON III (4k x 4k) |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.12_2829: / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||
| CTF correction | Type: NONE | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 178000 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C10 (10 fold cyclic) | ||||||||||||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 6.1 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 25000 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building |
| ||||||||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building |
| ||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller





 PDBj
PDBj