+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3885 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
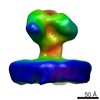
| Title | YaxAB pore complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Pore-forming toxin / Pathogens / Two-component toxin / MEMBRANE PROTEIN | |||||||||
| Function / homology | : / membrane => GO:0016020 / membrane / Uncharacterized protein / Membrane protein / Alpha-xenorhabdolysin family binary toxin subunit B Function and homology information Function and homology information | |||||||||
| Biological species |  Yersinia enterocolitica (bacteria) Yersinia enterocolitica (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.1 Å | |||||||||
 Authors Authors | Braeuning B / Bertosin E / Dietz H / Groll M | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: Structure and mechanism of the two-component α-helical pore-forming toxin YaxAB. Authors: Bastian Bräuning / Eva Bertosin / Florian Praetorius / Christian Ihling / Alexandra Schatt / Agnes Adler / Klaus Richter / Andrea Sinz / Hendrik Dietz / Michael Groll /  Abstract: Pore-forming toxins (PFT) are virulence factors that transform from soluble to membrane-bound states. The Yersinia YaxAB system represents a family of binary α-PFTs with orthologues in human, ...Pore-forming toxins (PFT) are virulence factors that transform from soluble to membrane-bound states. The Yersinia YaxAB system represents a family of binary α-PFTs with orthologues in human, insect, and plant pathogens, with unknown structures. YaxAB was shown to be cytotoxic and likely involved in pathogenesis, though the molecular basis for its two-component lytic mechanism remains elusive. Here, we present crystal structures of YaxA and YaxB, together with a cryo-electron microscopy map of the YaxAB complex. Our structures reveal a pore predominantly composed of decamers of YaxA-YaxB heterodimers. Both subunits bear membrane-active moieties, but only YaxA is capable of binding to membranes by itself. YaxB can subsequently be recruited to membrane-associated YaxA and induced to present its lytic transmembrane helices. Pore formation can progress by further oligomerization of YaxA-YaxB dimers. Our results allow for a comparison between pore assemblies belonging to the wider ClyA-like family of α-PFTs, highlighting diverse pore architectures. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3885.map.gz emd_3885.map.gz | 21.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3885-v30.xml emd-3885-v30.xml emd-3885.xml emd-3885.xml | 18.6 KB 18.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3885_fsc.xml emd_3885_fsc.xml | 14.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_3885.png emd_3885.png | 28.7 KB | ||
| Filedesc metadata |  emd-3885.cif.gz emd-3885.cif.gz | 6.2 KB | ||
| Others |  emd_3885_half_map_1.map.gz emd_3885_half_map_1.map.gz emd_3885_half_map_2.map.gz emd_3885_half_map_2.map.gz | 192 MB 192.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3885 http://ftp.pdbj.org/pub/emdb/structures/EMD-3885 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3885 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3885 | HTTPS FTP |
-Related structure data
| Related structure data |  6el1MC  6ek4C  6ek7C  6ek8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3885.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3885.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.106 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_3885_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_3885_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : YaxAB complex (10x YaxA + 10x YaxB)
| Entire | Name: YaxAB complex (10x YaxA + 10x YaxB) |
|---|---|
| Components |
|
-Supramolecule #1: YaxAB complex (10x YaxA + 10x YaxB)
| Supramolecule | Name: YaxAB complex (10x YaxA + 10x YaxB) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Purified with Cymal-6 detergent and reconstituted in amphipol prior to Cryo-EM. |
|---|---|
| Source (natural) | Organism:  Yersinia enterocolitica (bacteria) Yersinia enterocolitica (bacteria) |
| Molecular weight | Theoretical: 850 KDa |
-Macromolecule #1: YaxA
| Macromolecule | Name: YaxA / type: protein_or_peptide / ID: 1 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Yersinia enterocolitica (bacteria) Yersinia enterocolitica (bacteria) |
| Molecular weight | Theoretical: 45.877156 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TQTQLAIDNV LASAESTIQL NELPKVVLDF ITGEQTSVAR SGGIFTKEDL INLKLYVRKG LSLPTRQDEV EAYLGYKKID VAGLEPKDI KLLFDEIHNH ALNWNDVEQA VLQQSLDLDI AAKNIISTGN EIINLINQMP ITLRVKTLLG DITDKQLENI T YESADHEV ...String: TQTQLAIDNV LASAESTIQL NELPKVVLDF ITGEQTSVAR SGGIFTKEDL INLKLYVRKG LSLPTRQDEV EAYLGYKKID VAGLEPKDI KLLFDEIHNH ALNWNDVEQA VLQQSLDLDI AAKNIISTGN EIINLINQMP ITLRVKTLLG DITDKQLENI T YESADHEV ASALKDILDD MKGDINRHQT TTENVRKKVS DYRITLTGGE LSSGDKVNGL EPQVKTKYDL MEKSNMRKSI KE LDEKIKE KRQRIEQLKK DYDKFVGLSF TGAIGGIIAM AITGGIFGAK AENARKEKNA LISEVAELES KVSSQRALQT ALE ALSLSF SDIGIRMVDA ESALNHLDFM WLSVLNQITE SQIQFAMINN ALRLTSFVNK FQQVITPWQS VGDSARQLVD IFDE AIKEY KKVYG UniProtKB: Uncharacterized protein |
-Macromolecule #2: YaxB
| Macromolecule | Name: YaxB / type: protein_or_peptide / ID: 2 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Yersinia enterocolitica (bacteria) Yersinia enterocolitica (bacteria) |
| Molecular weight | Theoretical: 39.248836 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GAEISTFPHS GLSYPDINFK IFSQGVKNIS HLAQFKTTGV EVLQEKALRV SLYSQRLDVI VRESLSSLQV KLENTLALTY FTTLEEIDE ALISQDIDEE SKSEMRKERI NIIKNLSNDI TQLKQLFIEK TELLDKSSSD LHNVVIIEGT DKVLQAEQLR Q KQLTEDIA ...String: GAEISTFPHS GLSYPDINFK IFSQGVKNIS HLAQFKTTGV EVLQEKALRV SLYSQRLDVI VRESLSSLQV KLENTLALTY FTTLEEIDE ALISQDIDEE SKSEMRKERI NIIKNLSNDI TQLKQLFIEK TELLDKSSSD LHNVVIIEGT DKVLQAEQLR Q KQLTEDIA TKELERKEIE KKRDKIIEAL DVIREHNLVD AFKDLIPTGE NLSELDLAKP EIELLKQSLE ITKKLLGQFS EG LKYIDLT DARKKLDNQI DTASTRLTEL NRQLEQSEKL IAGVNAVIKI DQEKSAVVVE AEKLSRAWHI FIHEITALQG TSL NEVELS KPLIKQQIYL ESLIKQLI UniProtKB: Alpha-xenorhabdolysin family binary toxin subunit B |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
| |||||||||
| Grid | Model: C-flat-2/1 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: OTHER / Pretreatment - Pressure: 0.004 kPa | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 20 K / Instrument: FEI VITROBOT MARK IV Details: 3 mM F-FOS Choline 8 just before vitrification blot for 3 to 4 s blot distance -2 to -1 mm. | |||||||||
| Details | Sample exchanged to amphibole A8-35 and run in final round of gel filtration (buffer: 20 mM HEPES pH 7.0, 100 mM NaCl) prior to Cryo-EM. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: OTHER |
| Output model |  PDB-6el1: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)