[English] 日本語
 Yorodumi
Yorodumi- EMDB-9891: Cryo-EM structure of spike protein of feline infectious peritonit... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9891 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of spike protein of feline infectious peritonitis virus strain UU4 | ||||||||||||
 Map data Map data | FIPV-UU4 spike protein (sharpened) | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | CoV spike protein / VIRAL PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell endoplasmic reticulum-Golgi intermediate compartment membrane / receptor-mediated virion attachment to host cell / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion membrane / membrane Similarity search - Function | ||||||||||||
| Biological species |  Feline infectious peritonitis virus Feline infectious peritonitis virus | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.31 Å | ||||||||||||
 Authors Authors | Hsu STD / Yang TJ | ||||||||||||
| Funding support |  Taiwan, 3 items Taiwan, 3 items
| ||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2020 Journal: Proc Natl Acad Sci U S A / Year: 2020Title: Cryo-EM analysis of a feline coronavirus spike protein reveals a unique structure and camouflaging glycans. Authors: Tzu-Jing Yang / Yen-Chen Chang / Tzu-Ping Ko / Piotr Draczkowski / Yu-Chun Chien / Yuan-Chih Chang / Kuen-Phon Wu / Kay-Hooi Khoo / Hui-Wen Chang / Shang-Te Danny Hsu /  Abstract: Feline infectious peritonitis virus (FIPV) is an alphacoronavirus that causes a nearly 100% mortality rate without effective treatment. Here we report a 3.3-Å cryoelectron microscopy (cryo-EM) ...Feline infectious peritonitis virus (FIPV) is an alphacoronavirus that causes a nearly 100% mortality rate without effective treatment. Here we report a 3.3-Å cryoelectron microscopy (cryo-EM) structure of the serotype I FIPV spike (S) protein, which is responsible for host recognition and viral entry. Mass spectrometry provided site-specific compositions of densely distributed high-mannose and complex-type Nglycans that account for 1/4 of the total molecular mass; most of the N-glycans could be visualized by cryo-EM. Specifically, the N-glycans that wedge between 2 galectin-like domains within the S1 subunit of FIPV S protein result in a unique propeller-like conformation, underscoring the importance of glycosylation in maintaining protein structures. The cleavage site within the S2 subunit responsible for activation also showed distinct structural features and glycosylation. These structural insights provide a blueprint for a better molecular understanding of the pathogenesis of FIP. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9891.map.gz emd_9891.map.gz | 301.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9891-v30.xml emd-9891-v30.xml emd-9891.xml emd-9891.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9891.png emd_9891.png | 32.3 KB | ||
| Filedesc metadata |  emd-9891.cif.gz emd-9891.cif.gz | 6.9 KB | ||
| Others |  emd_9891_additional.map.gz emd_9891_additional.map.gz emd_9891_additional_1.map.gz emd_9891_additional_1.map.gz emd_9891_half_map_1.map.gz emd_9891_half_map_1.map.gz emd_9891_half_map_2.map.gz emd_9891_half_map_2.map.gz | 301.1 MB 301.1 MB 85 MB 85 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9891 http://ftp.pdbj.org/pub/emdb/structures/EMD-9891 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9891 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9891 | HTTPS FTP |
-Related structure data
| Related structure data |  6jx7MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9891.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9891.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | FIPV-UU4 spike protein (sharpened) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.87 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: FIPV-UU4 spike protein (unsharpened)
| File | emd_9891_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | FIPV-UU4 spike protein (unsharpened) | ||||||||||||
| Projections & Slices |
| ||||||||||||
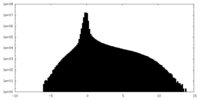
| Density Histograms |
-Additional map: FIPV-UU4 spike protein (unsharpened)
| File | emd_9891_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | FIPV-UU4 spike protein (unsharpened) | ||||||||||||
| Projections & Slices |
| ||||||||||||
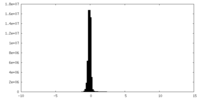
| Density Histograms |
-Half map: Half map 1
| File | emd_9891_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half_map_1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
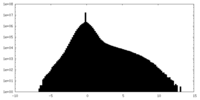
| Density Histograms |
-Half map: Half map 2
| File | emd_9891_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half_map_2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
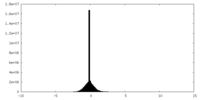
| Density Histograms |
- Sample components
Sample components
-Entire : Feline Infectious Peritonitis Virus Spike Protein
| Entire | Name: Feline Infectious Peritonitis Virus Spike Protein |
|---|---|
| Components |
|
-Supramolecule #1: Feline Infectious Peritonitis Virus Spike Protein
| Supramolecule | Name: Feline Infectious Peritonitis Virus Spike Protein / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Feline infectious peritonitis virus Feline infectious peritonitis virus |
| Molecular weight | Theoretical: 720 kDa/nm |
-Macromolecule #1: Feline Infectious Peritonitis Virus Spike Protein
| Macromolecule | Name: Feline Infectious Peritonitis Virus Spike Protein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Feline infectious peritonitis virus Feline infectious peritonitis virus |
| Molecular weight | Theoretical: 163.829844 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DAPHGVTLPH FNTSHNNSKF ELNFYNFLQT WDIPPNTETI LGGYLPYCDH EDNCGWYNFV YNNKVGPNAK YSYINTQNLN IPNVHGVYF DVREHNSDGV WDQIDRVGLL IAIHGTSHYS LLMVLQDGVE ASQPHVAVKI CHWNPGNIST YHQFDVNLGD G GQCVFNQR ...String: DAPHGVTLPH FNTSHNNSKF ELNFYNFLQT WDIPPNTETI LGGYLPYCDH EDNCGWYNFV YNNKVGPNAK YSYINTQNLN IPNVHGVYF DVREHNSDGV WDQIDRVGLL IAIHGTSHYS LLMVLQDGVE ASQPHVAVKI CHWNPGNIST YHQFDVNLGD G GQCVFNQR FSLDTVLTAN DFYGFQWTDT YVDIYLGGTI TKVWVVNDWS VVEASISSHW NALNYGYYIQ FVNRTTYYAY NS TGGSNYT HLQLTECHTD YCAGYAKNVF VPIDGKIPEG FSFSNWFLLT DKSTLVQGRV LSSQPVFVQC LRPVPTWSNN TAV VHFKND VFCPNVTADV LRFNLNFSDT DVYTDSTTDD QLHFTFEDNT TASITCYSSA NVTDNQPASG SISHTPFVSN SYLC FANFS HSSVSRQFLG ILPPTVREFA FGRDGSIFVN GYKYFSLQPI KSVNFSISSV ENYGFWTIAY TNYTDVMVDV NGTVI TRLF YCDSPLNRIK CQQLKHELPD GFYSASMLVK KDLPKTFVTM PQFYNWMNVT LHVVLNDIEK KADIILAGAP ELASLA DIH FEIAQANGSV VNVTSVCVQA RQLALFYKYT SLQGLYTYSN LVQLQNYDCP FSPQQFNNYL QFETLCFDVS PAVAGCK WS LVHDVKWRTQ FATITVSYKD GAMITTMPKA QLGFQDISNI VKDECTDYNI YGFQGTGIIR STTSRLVAGL YYTSASGD L LGFKISTTGE IFTVVPCDLT AQAAVINDEI VGAITATNQT DLFEFVNHTW SRSARGSSPS TVNTYTMPQF YYITKWNNG TSSNCTSVIT YSSFAICNTG EIKYVNVTHV EIVDDSVGVI KPVSTGNITI PKNFTVAVQA EYVQIQVKPV AVDCAKYVCN GNRHCLNLL TQYTSACQTI ENSLNLGARL ESLMLNDMIT VSDRSLEFAT VDKFNTTALG GEKLGGLYFD GLSSLLPPRV G MRSAVEDL LFNKVVTSGL GTVDDDYKKC SAGTDVADLV CAQYYNGIMV LPGVVDYNKM AMYTASLIGG MALGSITSAV AV PFSMQVQ ARLNYVALQT DVLQENQKIL ANAFNNAIGN ITLALGKVSN AITTVSDGFN SMASALTKIQ SVVNQQGEAL SHL ISQLQK NFQAISSSIA EIYNRLEKVE ADAQVDRLIT GRLAALNAYV AQTLTQYAEV KASRQLAMEK VNECVKSQSD RYGF CGNGT HLFSLVNSAP DGLLFFHTVL LPTEWEEVTA WSGICVNDTY AYLLKDFDHS IFSYNGTYMV TPRNMFQPRK PQMSD FVQI TSCEVTFLNT THTTFQEIVI DYIDINKTIA DMLEQYHSNY TTPELDLQLE IFNQTKLNLT AEIDQLEQRA DNLTNI AHE LQQYIDNLNK TIVDLEWLNR IETYVKWPGS GGYIPEAPRD GQAYVRKDGE WVLLSTFLKG QDNSADIQHS GGRSSLE GP RFEGKPIPNP LLGLDSTRTG HHHHHH |
-Macromolecule #9: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 9 / Number of copies: 27 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #10: alpha-D-mannopyranose
| Macromolecule | Name: alpha-D-mannopyranose / type: ligand / ID: 10 / Number of copies: 6 / Formula: MAN |
|---|---|
| Molecular weight | Theoretical: 180.156 Da |
| Chemical component information |  ChemComp-MAN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 1X D-PBS |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: blot for 3 seconds before plunging; blot force: 0; waiting time: 10s. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number real images: 2436 / Average exposure time: 2.5 sec. / Average electron dose: 1.5 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)