[日本語] English
 万見
万見- EMDB-9043: Yeast 26S proteasome bound to ubiquitinated substrate (5D motor state) -
+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-9043 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Yeast 26S proteasome bound to ubiquitinated substrate (5D motor state) | |||||||||
 マップデータ マップデータ | Yeast 26S proteasome bound to ubiquitinated substrate (5D motor state) | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | 26S Proteasome / ATPase / AAA+ / Protease / Motor protein / Ubiquitin | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報proteasome regulatory particle assembly / mitotic cell cycle phase transition / cytosolic proteasome complex / protein-containing complex localization / proteasome-activating activity / proteasome regulatory particle, base subcomplex / cyclin-dependent protein serine/threonine kinase regulator activity / nonfunctional rRNA decay / proteasome core complex assembly / nuclear outer membrane-endoplasmic reticulum membrane network ...proteasome regulatory particle assembly / mitotic cell cycle phase transition / cytosolic proteasome complex / protein-containing complex localization / proteasome-activating activity / proteasome regulatory particle, base subcomplex / cyclin-dependent protein serine/threonine kinase regulator activity / nonfunctional rRNA decay / proteasome core complex assembly / nuclear outer membrane-endoplasmic reticulum membrane network / Cross-presentation of soluble exogenous antigens (endosomes) / TNFR2 non-canonical NF-kB pathway / proteasomal ubiquitin-independent protein catabolic process / Ubiquitin Mediated Degradation of Phosphorylated Cdc25A / Regulation of PTEN stability and activity / peptide catabolic process / CDK-mediated phosphorylation and removal of Cdc6 / FBXL7 down-regulates AURKA during mitotic entry and in early mitosis / KEAP1-NFE2L2 pathway / Neddylation / Orc1 removal from chromatin / MAPK6/MAPK4 signaling / proteasome storage granule / Antigen processing: Ubiquitination & Proteasome degradation / positive regulation of RNA polymerase II transcription preinitiation complex assembly / proteasome core complex, alpha-subunit complex / Ub-specific processing proteases / ERAD pathway / Neutrophil degranulation / proteasome complex / nucleotide-excision repair / positive regulation of transcription elongation by RNA polymerase II / positive regulation of protein catabolic process / ubiquitin-dependent protein catabolic process / proteasome-mediated ubiquitin-dependent protein catabolic process / chromatin remodeling / protein domain specific binding / cell division / mRNA binding / ubiquitin protein ligase binding / ATP hydrolysis activity / mitochondrion / ATP binding / identical protein binding / nucleus / cytosol / cytoplasm 類似検索 - 分子機能 | |||||||||
| 生物種 |   Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
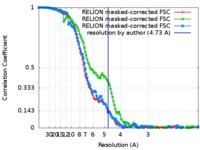
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 4.73 Å | |||||||||
 データ登録者 データ登録者 | de la Pena AH / Goodall EA | |||||||||
| 資金援助 |  米国, 2件 米国, 2件
| |||||||||
 引用 引用 |  ジャーナル: Science / 年: 2018 ジャーナル: Science / 年: 2018タイトル: Substrate-engaged 26 proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. 著者: Andres H de la Peña / Ellen A Goodall / Stephanie N Gates / Gabriel C Lander / Andreas Martin /  要旨: The 26 proteasome is the primary eukaryotic degradation machine and thus is critically involved in numerous cellular processes. The heterohexameric adenosine triphosphatase (ATPase) motor of the ...The 26 proteasome is the primary eukaryotic degradation machine and thus is critically involved in numerous cellular processes. The heterohexameric adenosine triphosphatase (ATPase) motor of the proteasome unfolds and translocates targeted protein substrates into the open gate of a proteolytic core while a proteasomal deubiquitinase concomitantly removes substrate-attached ubiquitin chains. However, the mechanisms by which ATP hydrolysis drives the conformational changes responsible for these processes have remained elusive. Here we present the cryo-electron microscopy structures of four distinct conformational states of the actively ATP-hydrolyzing, substrate-engaged 26 proteasome. These structures reveal how mechanical substrate translocation accelerates deubiquitination and how ATP-binding, -hydrolysis, and phosphate-release events are coordinated within the AAA+ (ATPases associated with diverse cellular activities) motor to induce conformational changes and propel the substrate through the central pore. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_9043.map.gz emd_9043.map.gz | 13.5 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-9043-v30.xml emd-9043-v30.xml emd-9043.xml emd-9043.xml | 56.5 KB 56.5 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_9043_fsc_1.xml emd_9043_fsc_1.xml emd_9043_fsc_2.xml emd_9043_fsc_2.xml emd_9043_fsc_3.xml emd_9043_fsc_3.xml | 12.2 KB 12.2 KB 12.1 KB | 表示 表示 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_9043.png emd_9043.png | 94.4 KB | ||
| マスクデータ |  emd_9043_msk_1.map emd_9043_msk_1.map emd_9043_msk_2.map emd_9043_msk_2.map emd_9043_msk_3.map emd_9043_msk_3.map | 149.9 MB 149.9 MB 149.9 MB |  マスクマップ マスクマップ | |
| Filedesc metadata |  emd-9043.cif.gz emd-9043.cif.gz | 10.1 KB | ||
| その他 |  emd_9043_additional_1.map.gz emd_9043_additional_1.map.gz emd_9043_additional_2.map.gz emd_9043_additional_2.map.gz emd_9043_additional_3.map.gz emd_9043_additional_3.map.gz emd_9043_additional_4.map.gz emd_9043_additional_4.map.gz emd_9043_additional_5.map.gz emd_9043_additional_5.map.gz emd_9043_additional_6.map.gz emd_9043_additional_6.map.gz emd_9043_additional_7.map.gz emd_9043_additional_7.map.gz emd_9043_additional_8.map.gz emd_9043_additional_8.map.gz emd_9043_additional_9.map.gz emd_9043_additional_9.map.gz | 118.3 MB 118.5 MB 140.2 MB 118.3 MB 118.3 MB 140.4 MB 118.4 MB 118.4 MB 140.4 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9043 http://ftp.pdbj.org/pub/emdb/structures/EMD-9043 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9043 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9043 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_9043.map.gz / 形式: CCP4 / 大きさ: 149.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_9043.map.gz / 形式: CCP4 / 大きさ: 149.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Yeast 26S proteasome bound to ubiquitinated substrate (5D motor state) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
+マスク #1
+マスク #2
+マスク #3
+追加マップ: Motor (half 2)
+追加マップ: Motor (half 1)
+追加マップ: Motor (sharpened)
+追加マップ: Alpha ring (half 2)
+追加マップ: Alpha ring (half 1)
+追加マップ: Alpha ring (sharpened)
+追加マップ: Global (half 2)
+追加マップ: Global (half 1)
+追加マップ: Global (sharpened)
- 試料の構成要素
試料の構成要素
+全体 : Substrate-engaged 26S proteasome in the 5D state (composite map)
+超分子 #1: Substrate-engaged 26S proteasome in the 5D state (composite map)
+超分子 #2: Proteasome
+超分子 #3: substrate
+分子 #1: Proteasome subunit alpha type-1
+分子 #2: Proteasome subunit alpha type-2
+分子 #3: Proteasome subunit alpha type-3
+分子 #4: Proteasome subunit alpha type-4
+分子 #5: Proteasome subunit alpha type-5
+分子 #6: Proteasome subunit alpha type-6
+分子 #7: Probable proteasome subunit alpha type-7
+分子 #8: 26S proteasome regulatory subunit 7 homolog
+分子 #9: 26S proteasome regulatory subunit 4 homolog
+分子 #10: 26S proteasome regulatory subunit 8 homolog
+分子 #11: 26S proteasome regulatory subunit 6B homolog
+分子 #12: 26S proteasome subunit RPT4
+分子 #13: 26S proteasome regulatory subunit 6A
+分子 #14: model substrate polypeptide
+分子 #15: ADENOSINE-5'-TRIPHOSPHATE
+分子 #16: ADENOSINE-5'-DIPHOSPHATE
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 25 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 緩衝液 | pH: 7.6 構成要素:
| ||||||||||||||||||
| グリッド | 支持フィルム - 材質: CARBON / 支持フィルム - トポロジー: HOLEY / 詳細: unspecified | ||||||||||||||||||
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 90 % / チャンバー内温度: 277 K / 装置: HOMEMADE PLUNGER 詳細: specimens were manually blotted with Whatman #1 filter paper. | ||||||||||||||||||
| 詳細 | 26S proteasomes were diluted to a concentration of 20 micromolar in a buffer with an ATP regeneration system, and 6 mM ortho-phenanthroline. This solution was mixed with an equal volume of 50 micromolar ubiquitinated model substrate |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 詳細 | images were acquired in nanoprobe mode |
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 検出モード: COUNTING / デジタル化 - サイズ - 横: 3710 pixel / デジタル化 - サイズ - 縦: 3838 pixel / デジタル化 - 画像ごとのフレーム数: 1-25 / 撮影したグリッド数: 1 / 実像数: 11656 / 平均露光時間: 6.25 sec. / 平均電子線量: 50.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | C2レンズ絞り径: 70.0 µm / 最大 デフォーカス(補正後): -3.0 µm / 最小 デフォーカス(補正後): -1.5 µm / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm / 最大 デフォーカス(公称値): -2.5 µm / 最小 デフォーカス(公称値): -1.0 µm / 倍率(公称値): 29000 |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー




































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)