[English] 日本語
 Yorodumi
Yorodumi- EMDB-8903: Cryo-EM Structures of ASC and NLRC4 CARD Filaments Reveal a Unifi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8903 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structures of ASC and NLRC4 CARD Filaments Reveal a Unified Mechanism of Nucleation and Activation of Caspase-1 | |||||||||
 Map data Map data | Helical reconstruction of NLRC4-CARD | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationIPAF inflammasome complex / The IPAF inflammasome / icosanoid biosynthetic process / canonical inflammasome complex / caspase binding / positive regulation of protein processing / pattern recognition receptor signaling pathway / TP53 Regulates Transcription of Caspase Activators and Caspases / pyroptotic inflammatory response / detection of bacterium ...IPAF inflammasome complex / The IPAF inflammasome / icosanoid biosynthetic process / canonical inflammasome complex / caspase binding / positive regulation of protein processing / pattern recognition receptor signaling pathway / TP53 Regulates Transcription of Caspase Activators and Caspases / pyroptotic inflammatory response / detection of bacterium / endopeptidase activator activity / activation of innate immune response / positive regulation of interleukin-1 beta production / : / protein homooligomerization / positive regulation of inflammatory response / defense response to bacterium / positive regulation of apoptotic process / inflammatory response / innate immune response / intracellular membrane-bounded organelle / apoptotic process / magnesium ion binding / protein homodimerization activity / ATP binding / identical protein binding / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.58 Å | |||||||||
 Authors Authors | Li Y / Fu T / Wu H | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2018 Journal: Proc Natl Acad Sci U S A / Year: 2018Title: Cryo-EM structures of ASC and NLRC4 CARD filaments reveal a unified mechanism of nucleation and activation of caspase-1. Authors: Yang Li / Tian-Min Fu / Alvin Lu / Kristen Witt / Jianbin Ruan / Chen Shen / Hao Wu /  Abstract: Canonical inflammasomes are cytosolic supramolecular complexes that activate caspase-1 upon sensing extrinsic microbial invasions and intrinsic sterile stress signals. During inflammasome assembly, ...Canonical inflammasomes are cytosolic supramolecular complexes that activate caspase-1 upon sensing extrinsic microbial invasions and intrinsic sterile stress signals. During inflammasome assembly, adaptor proteins ASC and NLRC4 recruit caspase-1 through homotypic caspase recruitment domain (CARD) interactions, leading to caspase-1 dimerization and activation. Activated caspase-1 processes proinflammatory cytokines and Gasdermin D to induce cytokine maturation and pyroptotic cell death. Here, we present cryo-electron microscopy (cryo-EM) structures of NLRC4 CARD and ASC CARD filaments mediated by conserved three types of asymmetric interactions (types I, II, and III). We find that the CARDs of these two adaptor proteins share a similar assembly pattern, which matches that of the caspase-1 CARD filament whose structure we defined previously. These data indicate a unified mechanism for downstream caspase-1 recruitment through CARD-CARD interactions by both adaptors. Using structure modeling, we further show that full-length NLRC4 assembles via two separate symmetries at its CARD and its nucleotide-binding domain (NBD), respectively. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8903.map.gz emd_8903.map.gz | 42.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8903-v30.xml emd-8903-v30.xml emd-8903.xml emd-8903.xml | 9.6 KB 9.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8903.png emd_8903.png | 78.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8903 http://ftp.pdbj.org/pub/emdb/structures/EMD-8903 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8903 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8903 | HTTPS FTP |
-Validation report
| Summary document |  emd_8903_validation.pdf.gz emd_8903_validation.pdf.gz | 533.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_8903_full_validation.pdf.gz emd_8903_full_validation.pdf.gz | 533.4 KB | Display | |
| Data in XML |  emd_8903_validation.xml.gz emd_8903_validation.xml.gz | 6.1 KB | Display | |
| Data in CIF |  emd_8903_validation.cif.gz emd_8903_validation.cif.gz | 6.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8903 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8903 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8903 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8903 | HTTPS FTP |
-Related structure data
| Related structure data |  6n1iMC  8902C  6n1hC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8903.map.gz / Format: CCP4 / Size: 45.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8903.map.gz / Format: CCP4 / Size: 45.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Helical reconstruction of NLRC4-CARD | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.32 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
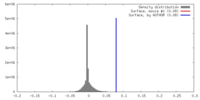
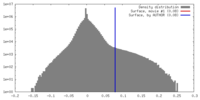
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Caspase recruitment domain of NLR family CARD domain-containing p...
| Entire | Name: Caspase recruitment domain of NLR family CARD domain-containing protein 4 |
|---|---|
| Components |
|
-Supramolecule #1: Caspase recruitment domain of NLR family CARD domain-containing p...
| Supramolecule | Name: Caspase recruitment domain of NLR family CARD domain-containing protein 4 type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism: |
-Macromolecule #1: NLR family CARD domain-containing protein 4
| Macromolecule | Name: NLR family CARD domain-containing protein 4 / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 9.987671 KDa |
| Recombinant expression | Organism: |
| Sequence | String: MNFIKDNSRA LIQRMGMTVI KQITDDLFVW NVLNREEVNI ICCEKVEQDA ARGIIHMILK KGSESCNLFL KSLKEWNYPL FQDLN |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 41.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 4.93 Å Applied symmetry - Helical parameters - Δ&Phi: -100.48 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 3.58 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 199312 |
|---|---|
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)