[English] 日本語
 Yorodumi
Yorodumi- EMDB-8073: Structure of open state rabbit ryanodine receptor 1 activated by ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8073 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of open state rabbit ryanodine receptor 1 activated by calcium ion | |||||||||
 Map data Map data | None | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationATP-gated ion channel activity / terminal cisterna / ryanodine receptor complex / ryanodine-sensitive calcium-release channel activity / release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / ossification involved in bone maturation / skin development / organelle membrane / cellular response to caffeine / outflow tract morphogenesis ...ATP-gated ion channel activity / terminal cisterna / ryanodine receptor complex / ryanodine-sensitive calcium-release channel activity / release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / ossification involved in bone maturation / skin development / organelle membrane / cellular response to caffeine / outflow tract morphogenesis / intracellularly gated calcium channel activity / toxic substance binding / voltage-gated calcium channel activity / smooth endoplasmic reticulum / skeletal muscle fiber development / striated muscle contraction / release of sequestered calcium ion into cytosol / muscle contraction / sarcoplasmic reticulum membrane / cellular response to calcium ion / sarcoplasmic reticulum / calcium ion transmembrane transport / calcium channel activity / sarcolemma / Z disc / intracellular calcium ion homeostasis / disordered domain specific binding / protein homotetramerization / transmembrane transporter binding / calmodulin binding / calcium ion binding / ATP binding / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.9 Å | |||||||||
 Authors Authors | Wang X / Wei R / Yin C / Sun F | |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2016 Journal: Cell Res / Year: 2016Title: Structural insights into Ca(2+)-activated long-range allosteric channel gating of RyR1. Authors: Risheng Wei / Xue Wang / Yan Zhang / Saptarshi Mukherjee / Lei Zhang / Qiang Chen / Xinrui Huang / Shan Jing / Congcong Liu / Shuang Li / Guangyu Wang / Yaofang Xu / Sujie Zhu / Alan J ...Authors: Risheng Wei / Xue Wang / Yan Zhang / Saptarshi Mukherjee / Lei Zhang / Qiang Chen / Xinrui Huang / Shan Jing / Congcong Liu / Shuang Li / Guangyu Wang / Yaofang Xu / Sujie Zhu / Alan J Williams / Fei Sun / Chang-Cheng Yin /   Abstract: Ryanodine receptors (RyRs) are a class of giant ion channels with molecular mass over 2.2 mega-Daltons. These channels mediate calcium signaling in a variety of cells. Since more than 80% of the RyR ...Ryanodine receptors (RyRs) are a class of giant ion channels with molecular mass over 2.2 mega-Daltons. These channels mediate calcium signaling in a variety of cells. Since more than 80% of the RyR protein is folded into the cytoplasmic assembly and the remaining residues form the transmembrane domain, it has been hypothesized that the activation and regulation of RyR channels occur through an as yet uncharacterized long-range allosteric mechanism. Here we report the characterization of a Ca(2+)-activated open-state RyR1 structure by cryo-electron microscopy. The structure has an overall resolution of 4.9 Å and a resolution of 4.2 Å for the core region. In comparison with the previously determined apo/closed-state structure, we observed long-range allosteric gating of the channel upon Ca(2+) activation. In-depth structural analyses elucidated a novel channel-gating mechanism and a novel ion selectivity mechanism of RyR1. Our work not only provides structural insights into the molecular mechanisms of channel gating and regulation of RyRs, but also sheds light on structural basis for channel-gating and ion selectivity mechanisms for the six-transmembrane-helix cation channel family. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8073.map.gz emd_8073.map.gz | 20.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8073-v30.xml emd-8073-v30.xml emd-8073.xml emd-8073.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8073.png emd_8073.png | 156.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8073 http://ftp.pdbj.org/pub/emdb/structures/EMD-8073 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8073 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8073 | HTTPS FTP |
-Validation report
| Summary document |  emd_8073_validation.pdf.gz emd_8073_validation.pdf.gz | 328.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_8073_full_validation.pdf.gz emd_8073_full_validation.pdf.gz | 327.9 KB | Display | |
| Data in XML |  emd_8073_validation.xml.gz emd_8073_validation.xml.gz | 7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8073 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8073 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8073 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8073 | HTTPS FTP |
-Related structure data
| Related structure data |  5j8vMC  8074C  8171C  8172C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8073.map.gz / Format: CCP4 / Size: 229.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8073.map.gz / Format: CCP4 / Size: 229.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.396 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
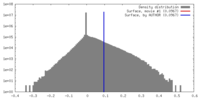
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Ryanodine Receptor
| Entire | Name: Ryanodine Receptor |
|---|---|
| Components |
|
-Supramolecule #1: Ryanodine Receptor
| Supramolecule | Name: Ryanodine Receptor / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.2 MDa |
-Macromolecule #1: ryanodine receptor
| Macromolecule | Name: ryanodine receptor / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MGDGGEGEDE VQFLRTDDEV VLQCSATVLK EQLKLCLAAE GFGNRLCFLE PTSNAQNVPP DLAICCFTLE QSLSVRALQE MLANTVEAGV ESSQGGGHRT LLYGHAILLR HAHSRMYLSC LTTSRSMTDK LAFDVGLQED ATGEACWWTM HPASKQRSEG EKVRVGDDLI ...String: MGDGGEGEDE VQFLRTDDEV VLQCSATVLK EQLKLCLAAE GFGNRLCFLE PTSNAQNVPP DLAICCFTLE QSLSVRALQE MLANTVEAGV ESSQGGGHRT LLYGHAILLR HAHSRMYLSC LTTSRSMTDK LAFDVGLQED ATGEACWWTM HPASKQRSEG EKVRVGDDLI LVSVSSERYL HLSTASGELQ VDASFMQTLW NMNPICSCCE EGYVTGGHVL RLFHGHMDEC LTISAADSDD QRRLVYYEGG AVCTHARSLW RLEPLRISWS GSHLRWGQPL RIRHVTTGRY LALTEDQGLV VVDACKAHTK ATSFCFRVSK EKLDTAPKRD VEGMGPPEIK YGESLCFVQH VASGLWLTYA APDPKALRLG VLKKKAILHQ EGHMDDALFL TRCQQEESQA ARMIHSTAGL YNQFIKGLDS FSGKPRGSGP PAGPALPIEA VILSLQDLIG YFEPPSEELQ HEEKQSKLRS LRNRQSLFQE EGMLSLVLNC IDRLNVYTTA AHFAEYAGEE AAESWKEIVN LLYELLASLI RGNRANCALF STNLDWVVSK LDRLEASSGI LEVLYCVLIE SPEVLNIIQE NHIKSIISLL DKHGRNHKVL DVLCSLCVCN GVAVRSNQDL ITENLLPGRE LLLQTNLINY VTSIRPNIFV GRAEGSTQYG KWYFEVMVDE VVPFLTAQAT HLRVGWALTE GYSPYPGGGE GWGGNGVGDD LYSYGFDGLH LWTGHVARPV TSPGQHLLAP EDVVSCCLDL SVPSISFRIN GCPVQGVFEA FNLDGLFFPV VSFSAGVKVR FLLGGRHGEF KFLPPPGYAP CHEAVLPRER LRLEPIKEYR REGPRGPHLV GPSRCLSHTD FVPCPVDTVQ IVLPPHLERI REKLAENIHE LWALTRIEQG WTYGPVRDDN KRLHPCLVNF HSLPEPERNY NLQMSGETLK TLLALGCHVG MADEKAEDNL KKTKLPKTYM MSNGYKPAPL DLSHVRLTPA QTTLVDRLAE NGHNVWARDR VAQGWSYSAV QDIPARRNPR LVPYRLLDEA TKRSNRDSLC QAVRTLLGYG YNIEPPDQEP SQVENQSRWD RVRIFRAEKS YTVQSGRWYF EFEAVTTGEM RVGWARPELR PDVELGADEL AYVFNGHRGQ RWHLGSEPFG RPWQSGDVVG CMIDLTENTI IFTLNGEVLM SDSGSETAFR EIEIGDGFLP VCSLGPGQVG HLNLGQDVSS LRFFAICGLQ EGFEPFAINM QRPVTTWFSK SLPQFEPVPP EHPHYEVARM DGTVDTPPCL RLAHRTWGSQ NSLVEMLFLR LSLPVQFHQH FRCTAGATPL APPGLQPPAE DEARAAEPDP DYENLRRSAG GWGEAEGGKE GTAKEGTPGG TPQPGVEAQP VRAENEKDAT TEKNKKRGFL FKAKKAAMMT QPPATPALPR LPHDVVPADN RDDPEIILNT TTYYYSVRVF AGQEPSCVWV GWVTPDYHQH DMNFDLSKVR AVTVTMGDEQ GNVHSSLKCS NCYMVWGGDF VSPGQQGRIS HTDLVIGCLV DLATGLMTFT ANGKESNTFF QVEPNTKLFP AVFVLPTHQN VIQFELGKQK NIMPLSAAMF LSERKNPAPQ CPPRLEVQML MPVSWSRMPN HFLQVETRRA GERLGWAVQC QDPLTMMALH IPEENRCMDI LELSERLDLQ RFHSHTLRLY RAVCALGNNR VAHALCSHVD QAQLLHALED AHLPGPLRAG YYDLLISIHL ESACRSRRSM LSEYIVPLTP ETRAITLFPP GRKGGNARRH GLPGVGVTTS LRPPHHFSPP CFVAALPAAG VAEAPARLSP AIPLEALRDK ALRMLGEAVR DGGQHARDPV GGSVEFQFVP VLKLVSTLLV MGIFGDEDVK QILKMIEPEV FTEEEEEEEE EEEEEEEEEE DEEEKEEDEE EEEKEDAEKE EEEAPEGEKE DLEEGLLQMK LPESVKLQMC NLLEYFCDQE LQHRVESLAA FAERYVDKLQ ANQRSRYALL MRAFTMSAAE TARRTREFRS PPQEQINMLL HFKDEADEED CPLPEDIRQD LQDFHQDLLA HCGIQLEGEE EEPEEETSLS SRLRSLLETV RLVKKKEEKP EEELPAEEKK PQSLQELVSH MVVRWAQEDY VQSPELVRAM FSLLHRQYDG LGELLRALPR AYTISPSSVE DTMSLLECLG QIRSLLIVQM GPQEENLMIQ SIGNIMNNKV FYQHPNLMRA LGMHETVMEV MVNVLGGGET KEIRFPKMVT SCCRFLCYFC RISRQNQRSM FDHLSYLLEN SGIGLGMQGS TPLDVAAASV IDNNELALAL QEQDLEKVVS YLAGCGLQSC PMLLAKGYPD IGWNPCGGER YLDFLRFAVF VNGESVEENA NVVVRLLIRK PECFGPALRG EGGSGLLAAI EEAIRISEDP ARDGPGVRRD RRREHFGEEP PEENRVHLGH AIMSFYAALI DLLGRCAPEM HLIQAGKGEA LRIRAILRSL VPLDDLVGII SLPLQIPTLG KDGALVQPKM SASFVPDHKA SMVLFLDRVY GIENQDFLLH VLDVGFLPDM RAAASLDTAT FSTTEMALAL NRYLCLAVLP LITKCAPLFA GTEHRAIMVD SMLHTVYRLS RGRSLTKAQR DVIEDCLMAL CRYIRPSMLQ HLLRRLVFDV PILNEFAKMP LKLLTNHYER CWKYYCLPTG WANFGVTSEE ELHLTRKLFW GIFDSLAHKK YDQELYRMAM PCLCAIAGAL PPDYVDASYS SKAEKKATVD AEGNFDPRPV ETLNVIIPEK LDSFINKFAE YTHEKWAFDK IQNNWSYGEN VDEELKTHPM LRPYKTFSEK DKEIYRWPIK ESLKAMIAWE WTIEKAREGE EERTEKKKTR KISQTAQTYD PREGYNPQPP DLSGVTLSRE LQAMAEQLAE NYHNTWGRKK KQELEAKGGG THPLLVPYDT LTAKEKARDR EKAQELLKFL QMNGYAVTRG LKDMELDTSS IEKRFAFGFL QQLLRWMDIS QEFIAHLEAV VSSGRVEKSP HEQEIKFFAK ILLPLINQYF TNHCLYFLST PAKVLGSGGH ASNKEKEMIT SLFCKLAALV RHRVSLFGTD APAVVNCLHI LARSLDARTV MKSGPEIVKA GLRSFFESAS EDIEKMVENL RLGKVSQART QVKGVGQNLT YTTVALLPVL TTLFQHIAQH QFGDDVILDD VQVSCYRTLC SIYSLGTTKN TYVEKLRPAL GECLARLAAA MPVAFLEPQL NEYNACSVYT TKSPRERAIL GLPNSVEEMC PDIPVLDRLM ADIGGLAESG ARYTEMPHVI EITLPMLCSY LPRWWERGPE APPPALPAGA PPPCTAVTSD HLNSLLGNIL RIIVNNLGID EATWMKRLAV FAQPIVSRAR PELLHSHFIP TIGRLRKRAG KVVAEEEQLR LEAKAEAEEG ELLVRDEFSV LCRDLYALYP LLIRYVDNNR AHWLTEPNAN AEELFRMVGE IFIYWSKSHN FKREEQNFVV QNEINNMSFL TADSKSKMAK AGDAQSGGSD QERTKKKRRG DRYSVQTSLI VATLKKMLPI GLNMCAPTDQ DLIMLAKTRY ALKDTDEEVR EFLQNNLHLQ GKVEGSPSLR WQMALYRGLP GREEDADDPE KIVRRVQEVS AVLYHLEQTE HPYKSKKAVW HKLLSKQRRR AVVACFRMTP LYNLPTHRAC NMFLESYKAA WILTEDHSFE DRMIDDLSKA GEQEEEEEEV EEKKPDPLHQ LVLHFSRTAL TEKSKLDEDY LYMAYADIMA KSCHLEEGGE NGEAEEEEVE VSFEEKEMEK QRLLYQQSRL HTRGAAEMVL QMISACKGET GAMVSSTLKL GISILNGGNA EVQQKMLDYL KDKKEVGFFQ SIQALMQTCS VLDLNAFERQ NKAEGLGMVN EDGTVINRQN GEKVMADDEF TQDLFRFLQL LCEGHNNDFQ NYLRTQTGNT TTINIIICTV DYLLRLQESI SDFYWYYSGK DVIEEQGKRN FSKAMSVAKQ VFNSLTEYIQ GPCTGNQQSL AHSRLWDAVV GFLHVFAHMM MKLAQDSSQI ELLKELLDLQ KDMVVMLLSL LEGNVVNGMI ARQMVDMLVE SSSNVEMILK FFDMFLKLKD IVGSEAFQDY VTDPRGLISK KDFQKAMDSQ KQFTGPEIQF LLSCSEADEN EMINFEEFAN RFQEPARDIG FNVAVLLTNL SEHVPHDPRL RNFLELAESI LEYFRPYLGR IEIMGASRRI ERIYFEISET NRAQWEMPQV KESKRQFIFD VVNEGGEAEK MELFVSFCED TIFEMQIAAQ ISEPEGEPEA DEDEGMGEAA AEGAEEGAAG AEGAAGTVAA GATARLAAAA ARALRGLSYR SLRRRVRRLR RLTAREAATA LAALLWAVVA RAGAAGAGAA AGALRLLWGS LFGGGLVEGA KKVTVTELLA GMPDPTSDEV HGEQPAGPGG DADGAGEGEG EGDAAEGDGD EEVAGHEAGP GGAEGVVAVA DGGPFRPEGA GGLGDMGDTT PAEPPTPEGS PILKRKLGVD GEEEELVPEP EPEPEPEPEK ADEENGEKEE VPEAPPEPPK KAPPSPPAKK EEAGGAGMEF WGELEVQRVK FLNYLSRNFY TLRFLALFLA FAINFILLFY KVSDSPPGED DMEGSAAGDL AGAGSGGGSG WGSGAGEEAE GDEDENMVYY FLEESTGYME PALWCLSLLH TLVAFLCIIG YNCLKVPLVI FKREKELARK LEFDGLYITE QPGDDDVKGQ WDRLVLNTPS FPSNYWDKFV KRKVLDKHGD IFGRERIAEL LGMDLASLEI TAHNERKPDP PPGLLTWLMS IDVKYQIWKF GVIFTDNSFL YLGWYMVMSL LGHYNNFFFA AHLLDIAMGV KTLRTILSSV THNGKQLVMT VGLLAVVVYL YTVVAFNFFR KFYNKSEDED EPDMKCDDMM TCYLFHMYVG VRAGGGIGDE IEDPAGDEYE LYRVVFDITF FFFVIVILLA IIQGLIIDAF GELRDQQEQV KEDMETKCFI CGIGSDYFDT TPHGFETHTL EEHNLANYMF FLMYLINKDE TEHTGQESYV WKMYQERCWD FFPAGDCFRK QYEDQLS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: 1:1000 diluted protease inhibitor cocktail was also added in buffer. | |||||||||||||||
| Grid | Model: Quantifoil R2.0/2.0 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: OTHER | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: Grids were blotted for 2s before plunging.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 80.0 K / Max: 80.0 K |
| Alignment procedure | Coma free - Residual tilt: 23.0 mrad |
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Sampling interval: 14.0 µm / Digitization - Frames/image: 2-31 / Number grids imaged: 4 / Number real images: 4931 / Average exposure time: 2.0 sec. / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 5.4 µm / Calibrated defocus min: 1.3 µm / Calibrated magnification: 100286 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)