[English] 日本語
 Yorodumi
Yorodumi- EMDB-7132: Human TRPM4 ion channel in lipid nanodiscs in a calcium-free state -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7132 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human TRPM4 ion channel in lipid nanodiscs in a calcium-free state | ||||||||||||||||||
 Map data Map data | Sharpened 3D density map of human TRPM4 in lipid nanodisc. This reconstruction includes the whole molecule. | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | TRPM4 / TRPM channel / TRP channel / MEMBRANE PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of atrial cardiac muscle cell action potential / positive regulation of regulation of vascular associated smooth muscle cell membrane depolarization / sodium channel complex / regulation of T cell cytokine production / membrane depolarization during AV node cell action potential / metal ion transport / membrane depolarization during bundle of His cell action potential / membrane depolarization during Purkinje myocyte cell action potential / negative regulation of bone mineralization / regulation of ventricular cardiac muscle cell action potential ...positive regulation of atrial cardiac muscle cell action potential / positive regulation of regulation of vascular associated smooth muscle cell membrane depolarization / sodium channel complex / regulation of T cell cytokine production / membrane depolarization during AV node cell action potential / metal ion transport / membrane depolarization during bundle of His cell action potential / membrane depolarization during Purkinje myocyte cell action potential / negative regulation of bone mineralization / regulation of ventricular cardiac muscle cell action potential / calcium-activated cation channel activity / sodium ion import across plasma membrane / inorganic cation transmembrane transport / dendritic cell chemotaxis / TRP channels / cellular response to ATP / sodium channel activity / monoatomic cation transmembrane transport / regulation of heart rate by cardiac conduction / positive regulation of insulin secretion involved in cellular response to glucose stimulus / protein sumoylation / negative regulation of osteoblast differentiation / positive regulation of fat cell differentiation / positive regulation of heart rate / positive regulation of adipose tissue development / positive regulation of vasoconstriction / calcium-mediated signaling / calcium ion transmembrane transport / calcium channel activity / Sensory perception of sweet, bitter, and umami (glutamate) taste / positive regulation of canonical Wnt signaling pathway / positive regulation of cytosolic calcium ion concentration / protein homotetramerization / adaptive immune response / calmodulin binding / neuronal cell body / positive regulation of cell population proliferation / calcium ion binding / endoplasmic reticulum / Golgi apparatus / nucleoplasm / ATP binding / identical protein binding / membrane / plasma membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | ||||||||||||||||||
 Authors Authors | Autzen HE / Myasnikov AG | ||||||||||||||||||
| Funding support |  United States, United States,  Denmark, 5 items Denmark, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Science / Year: 2018 Journal: Science / Year: 2018Title: Structure of the human TRPM4 ion channel in a lipid nanodisc. Authors: Henriette E Autzen / Alexander G Myasnikov / Melody G Campbell / Daniel Asarnow / David Julius / Yifan Cheng /   Abstract: Transient receptor potential (TRP) melastatin 4 (TRPM4) is a widely expressed cation channel associated with a variety of cardiovascular disorders. TRPM4 is activated by increased intracellular ...Transient receptor potential (TRP) melastatin 4 (TRPM4) is a widely expressed cation channel associated with a variety of cardiovascular disorders. TRPM4 is activated by increased intracellular calcium in a voltage-dependent manner but, unlike many other TRP channels, is permeable to monovalent cations only. Here we present two structures of full-length human TRPM4 embedded in lipid nanodiscs at ~3-angstrom resolution, as determined by single-particle cryo-electron microscopy. These structures, with and without calcium bound, reveal a general architecture for this major subfamily of TRP channels and a well-defined calcium-binding site within the intracellular side of the S1-S4 domain. The structures correspond to two distinct closed states. Calcium binding induces conformational changes that likely prime the channel for voltage-dependent opening. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7132.map.gz emd_7132.map.gz | 5.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7132-v30.xml emd-7132-v30.xml emd-7132.xml emd-7132.xml | 29.3 KB 29.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7132.png emd_7132.png | 191.3 KB | ||
| Filedesc metadata |  emd-7132.cif.gz emd-7132.cif.gz | 6.9 KB | ||
| Others |  emd_7132_additional_1.map.gz emd_7132_additional_1.map.gz emd_7132_additional_2.map.gz emd_7132_additional_2.map.gz emd_7132_additional_3.map.gz emd_7132_additional_3.map.gz emd_7132_additional_4.map.gz emd_7132_additional_4.map.gz emd_7132_half_map_1.map.gz emd_7132_half_map_1.map.gz emd_7132_half_map_2.map.gz emd_7132_half_map_2.map.gz | 93.9 MB 9.1 MB 3.2 MB 9.9 MB 94 MB 93.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7132 http://ftp.pdbj.org/pub/emdb/structures/EMD-7132 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7132 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7132 | HTTPS FTP |
-Related structure data
| Related structure data |  6bqrMC  7133C  6bqvC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10126 (Title: Human TRPM4 ion channel in a lipid nanodisc in a calcium-free state EMPIAR-10126 (Title: Human TRPM4 ion channel in a lipid nanodisc in a calcium-free stateData size: 162.5 Data #1: particle stacks of TRPM4 particles post 2D clean-up [picked particles - multiframe - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7132.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7132.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened 3D density map of human TRPM4 in lipid nanodisc. This reconstruction includes the whole molecule. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
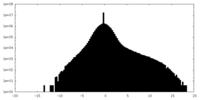
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.046 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Unsharpened 3D density map of human TRPM4 in...
| File | emd_7132_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened 3D density map of human TRPM4 in lipid nanodisc. The reconstruction includes the whole molecule. | ||||||||||||
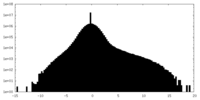
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sharpened 3D density map of human TRPM4 in...
| File | emd_7132_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened 3D density map of human TRPM4 in lipid nanodisc. This reconstruction includes only the cytoplasmic domain refined as a monomer, showing connectivity but fewer high-resolution features. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sharpened 3D density map of human TRPM4 in...
| File | emd_7132_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened 3D density map of human TRPM4 in lipid nanodisc. The reconstruction includes only the transmembrane domain and part of the cytoplasmic domain, refined with C4 symmetry | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sharpened 3D density map of human TRPM4 in...
| File | emd_7132_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened 3D density map of human TRPM4 in lipid nanodisc. This reconstruction includes only the cytoplasmic domain refined as a monomer, showing high-resolution features but less connectivity. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unsharpened 3D density half map #1 of human...
| File | emd_7132_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened 3D density half map #1 of human TRPM4 in lipid nanodisc. The reconstruction includes the whole molecule. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unsharpened 3D density half map #2 of human...
| File | emd_7132_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened 3D density half map #2 of human TRPM4 in lipid nanodisc. The reconstruction includes the whole molecule. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human TRPM4 ion channel
| Entire | Name: Human TRPM4 ion channel |
|---|---|
| Components |
|
-Supramolecule #1: Human TRPM4 ion channel
| Supramolecule | Name: Human TRPM4 ion channel / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Human TRPM4 ion channel in lipid nanodiscs in a calcium-free state |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 134.6 KDa |
-Macromolecule #1: Transient receptor potential cation channel subfamily M member 4
| Macromolecule | Name: Transient receptor potential cation channel subfamily M member 4 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 121.477953 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: YGELDFTGAG RKHSNFLRLS DRTDPAAVYS LVTRTWGFRA PNLVVSVLGG SGGPVLQTWL QDLLRRGLVR AAQSTGAWIV TGGLHTGIG RHVGVAVRDH QMASTGGTKV VAMGVAPWGV VRNRDTLINP KGSFPARYRW RGDPEDGVQF PLDYNYSAFF L VDDGTHGC ...String: YGELDFTGAG RKHSNFLRLS DRTDPAAVYS LVTRTWGFRA PNLVVSVLGG SGGPVLQTWL QDLLRRGLVR AAQSTGAWIV TGGLHTGIG RHVGVAVRDH QMASTGGTKV VAMGVAPWGV VRNRDTLINP KGSFPARYRW RGDPEDGVQF PLDYNYSAFF L VDDGTHGC LGGENRFRLR LESYISQQKT GVGGTGIDIP VLLLLIDGDE KMLTRIENAT QAQLPCLLVA GSGGAADCLA ET LEDTLAP GSGGARQGEA RDRIRRFFPK GDLEVLQAQV ERIMTRKELL TVYSSEDGSE EFETIVLKAL VKACGSSEAS AYL DELRLA VAWNRVDIAQ SELFRGDIQW RSFHLEASLM DALLNDRPEF VRLLISHGLS LGHFLTPMRL AQLYSAAPSN SLIR NLLDQ ASHSAGTKAP ALKGGAAELR PPDVGHVLRM LLGKMCAPRY PSGGAWDPHP GQGFGESMYL LSDKATSPLS LDAGL GQAP WSDLLLWALL LNRAQMAMYF WEMGSNAVSS ALGACLLLRV MARLEPDAEE AARRKDLAFK FEGMGVDLFG ECYRSS EVR AARLLLRRCP LWGDATCLQL AMQADARAFF AQDGVQSLLT QKWWGDMAST TPIWALVLAF FCPPLIYTRL ITFRKSE EE PTREELEFDM DSVINGEGPV GTADPAEKTP LGVPRQSGRP GCCGGRCGGR RCLRRWFHFW GAPVTIFMGN VVSYLLFL L LFSRVLLVDF QPAPPGSLEL LLYFWAFTLL CEELRQGLSG GGGSLASGGP GPGHASLSQR LRLYLADSWN QCDLVALTC FLLGVGCRLT PGLYHLGRTV LCIDFMVFTV RLLHIFTVNK QLGPKIVIVS KMMKDVFFFL FFLGVWLVAY GVATEGLLRP RDSDFPSIL RRVFYRPYLQ IFGQIPQEDM DVALMEHSNC SSEPGFWAHP PGAQAGTCVS QYANWLVVLL LVIFLLVANI L LVNLLIAM FSYTFGKVQG NSDLYWKAQR YRLIREFHSR PALAPPFIVI SHLRLLLRQL CRRPRSPQPS SPALEHFRVY LS KEAERKL LTWESVHKEN FLLARARDKR ESDSERLKRT SQKVDLALKQ LGHIREY UniProtKB: Transient receptor potential cation channel subfamily M member 4 |
-Macromolecule #2: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 2 / Number of copies: 12 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Number real images: 1817 / Average exposure time: 10.0 sec. / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6bqr: |
 Movie
Movie Controller
Controller





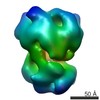
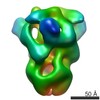



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)