登録情報 データベース : EMDB / ID : EMD-7109タイトル S. cerevisiae spliceosomal post-catalytic P complex Spliceosomal post-catalytic P complex 複合体 : Spliceosomal P complexRNA : x 5種タンパク質・ペプチド : x 31種リガンド : x 4種機能・相同性 分子機能 ドメイン・相同性 構成要素
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Saccharomyces cerevisiae (パン酵母) / Baker's yeast (パン酵母)手法 / / 解像度 : 3.3 Å Liu S / Li X / Zhou ZH / Zhao R 資金援助 Organization Grant number 国 National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) GM114178 National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) AI094386 National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) GM071940
ジャーナル : Science / 年 : 2017タイトル : Structure of the yeast spliceosomal postcatalytic P complex.著者 : Shiheng Liu / Xueni Li / Lingdi Zhang / Jiansen Jiang / Ryan C Hill / Yanxiang Cui / Kirk C Hansen / Z Hong Zhou / Rui Zhao / 要旨 : The spliceosome undergoes dramatic changes in a splicing cycle. Structures of B, B, C, C*, and intron lariat spliceosome complexes revealed mechanisms of 5'-splice site (ss) recognition, branching, ... The spliceosome undergoes dramatic changes in a splicing cycle. Structures of B, B, C, C*, and intron lariat spliceosome complexes revealed mechanisms of 5'-splice site (ss) recognition, branching, and intron release, but lacked information on 3'-ss recognition, exon ligation, and exon release. Here we report a cryo-electron microscopy structure of the postcatalytic P complex at 3.3-angstrom resolution, revealing that the 3' ss is mainly recognized through non-Watson-Crick base pairing with the 5' ss and branch point. Furthermore, one or more unidentified proteins become stably associated with the P complex, securing the 3' exon and potentially regulating activity of the helicase Prp22. Prp22 binds nucleotides 15 to 21 in the 3' exon, enabling it to pull the intron-exon or ligated exons in a 3' to 5' direction to achieve 3'-ss proofreading or exon release, respectively. 履歴 登録 2017年11月7日 - ヘッダ(付随情報) 公開 2018年2月21日 - マップ公開 2018年2月21日 - 更新 2020年10月14日 - 現状 2020年10月14日 処理サイト : RCSB / 状態 : 公開
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報 マップデータ
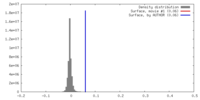
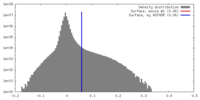
マップデータ 試料
試料 機能・相同性情報
機能・相同性情報

 データ登録者
データ登録者 米国, 3件
米国, 3件  引用
引用 ジャーナル: Science / 年: 2017
ジャーナル: Science / 年: 2017
 構造の表示
構造の表示 ムービービューア
ムービービューア SurfView
SurfView Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク emd_7109.map.gz
emd_7109.map.gz EMDBマップデータ形式
EMDBマップデータ形式 emd-7109-v30.xml
emd-7109-v30.xml emd-7109.xml
emd-7109.xml EMDBヘッダ
EMDBヘッダ emd_7109.png
emd_7109.png http://ftp.pdbj.org/pub/emdb/structures/EMD-7109
http://ftp.pdbj.org/pub/emdb/structures/EMD-7109 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7109
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7109 emd_7109_validation.pdf.gz
emd_7109_validation.pdf.gz EMDB検証レポート
EMDB検証レポート emd_7109_full_validation.pdf.gz
emd_7109_full_validation.pdf.gz emd_7109_validation.xml.gz
emd_7109_validation.xml.gz emd_7109_validation.cif.gz
emd_7109_validation.cif.gz https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7109
https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7109 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7109
ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7109 リンク
リンク EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource マップ
マップ ダウンロード / ファイル: emd_7109.map.gz / 形式: CCP4 / 大きさ: 244.1 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
ダウンロード / ファイル: emd_7109.map.gz / 形式: CCP4 / 大きさ: 244.1 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) 試料の構成要素
試料の構成要素 解析
解析 試料調製
試料調製 電子顕微鏡法
電子顕微鏡法 FIELD EMISSION GUN
FIELD EMISSION GUN
 画像解析
画像解析
 ムービー
ムービー コントローラー
コントローラー





























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)