+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6975 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 3.3 angstrom structure of mouse TRPM7 with EDTA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CryoEM / Truncated mouse TRPM7 / MEMBRANE PROTEIN / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationcalcium-dependent cell-matrix adhesion / intracellular magnesium ion homeostasis / magnesium ion transmembrane transport / zinc ion transport / zinc ion transmembrane transporter activity / magnesium ion transmembrane transporter activity / TRP channels / actomyosin structure organization / myosin binding / necroptotic process ...calcium-dependent cell-matrix adhesion / intracellular magnesium ion homeostasis / magnesium ion transmembrane transport / zinc ion transport / zinc ion transmembrane transporter activity / magnesium ion transmembrane transporter activity / TRP channels / actomyosin structure organization / myosin binding / necroptotic process / ruffle / cytoplasmic vesicle membrane / calcium channel activity / kinase activity / calcium ion transport / protein autophosphorylation / actin binding / cytoplasmic vesicle / protein homotetramerization / protein kinase activity / non-specific serine/threonine protein kinase / protein serine kinase activity / protein serine/threonine kinase activity / ATP binding / metal ion binding / nucleus / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
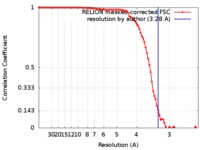
| Method | single particle reconstruction / cryo EM / Resolution: 3.28 Å | |||||||||
 Authors Authors | Zhang J / Li Z | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2018 Journal: Proc Natl Acad Sci U S A / Year: 2018Title: Structure of the mammalian TRPM7, a magnesium channel required during embryonic development. Authors: Jingjing Duan / Zongli Li / Jian Li / Raymond E Hulse / Ana Santa-Cruz / William C Valinsky / Sunday A Abiria / Grigory Krapivinsky / Jin Zhang / David E Clapham /   Abstract: The transient receptor potential ion channel subfamily M, member 7 (TRPM7), is a ubiquitously expressed protein that is required for mouse embryonic development. TRPM7 contains both an ion channel ...The transient receptor potential ion channel subfamily M, member 7 (TRPM7), is a ubiquitously expressed protein that is required for mouse embryonic development. TRPM7 contains both an ion channel and an α-kinase. The channel domain comprises a nonselective cation channel with notable permeability to Mg and Zn Here, we report the closed state structures of the mouse TRPM7 channel domain in three different ionic conditions to overall resolutions of 3.3, 3.7, and 4.1 Å. The structures reveal key residues for an ion binding site in the selectivity filter, with proposed partially hydrated Mg ions occupying the center of the conduction pore. In high [Mg], a prominent external disulfide bond is found in the pore helix, which is essential for ion channel function. Our results provide a structural framework for understanding the TRPM1/3/6/7 subfamily and extend the knowledge base upon which to study the diversity and evolution of TRP channels. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6975.map.gz emd_6975.map.gz | 6.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6975-v30.xml emd-6975-v30.xml emd-6975.xml emd-6975.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6975_fsc.xml emd_6975_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_6975.png emd_6975.png | 189.5 KB | ||
| Filedesc metadata |  emd-6975.cif.gz emd-6975.cif.gz | 7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6975 http://ftp.pdbj.org/pub/emdb/structures/EMD-6975 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6975 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6975 | HTTPS FTP |
-Related structure data
| Related structure data |  5zx5MC  7297C  7298C  6bwdC  6bwfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6975.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6975.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.23 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Membrane Protein, Transient Receptor Potential ion channel
| Entire | Name: Membrane Protein, Transient Receptor Potential ion channel |
|---|---|
| Components |
|
-Supramolecule #1: Membrane Protein, Transient Receptor Potential ion channel
| Supramolecule | Name: Membrane Protein, Transient Receptor Potential ion channel type: cell / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Transient receptor potential cation channel subfamily M member 7
| Macromolecule | Name: Transient receptor potential cation channel subfamily M member 7 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 141.417719 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SQKSWIESTL TKRECVYIIP SSKDPHRCLP GCQICQQLVR CFCGRLVKQH ACFTASLAMK YSDVKLGEHF NQAIEEWSVE KHTEQSPTD AYGVINFQGG SHSYRAKYVR LSYDTKPEII LQLLLKEWQM ELPKLVISVH GGMQKFELHP RIKQLLGKGL I KAAVTTGA ...String: SQKSWIESTL TKRECVYIIP SSKDPHRCLP GCQICQQLVR CFCGRLVKQH ACFTASLAMK YSDVKLGEHF NQAIEEWSVE KHTEQSPTD AYGVINFQGG SHSYRAKYVR LSYDTKPEII LQLLLKEWQM ELPKLVISVH GGMQKFELHP RIKQLLGKGL I KAAVTTGA WILTGGVNTG VAKHVGDALK EHASRSSRKI CTIGIAPWGV IENRNDLVGR DVVAPYQTLL NPLSKLNVLN NL HSHFILV DDGTVGKYGA EVRLRRELEK TINQQRIHAR IGQGVPVVAL IFEGGPNVIL TVLEYLQESP PVPVVVCEGT GRA ADLLAY IHKQTEEGGN LPDAAEPDII STIKKTFNFG QSEAVHLFQT MMECMKKKEL ITVFHIGSED HQDIDVAILT ALLK GTNAS AFDQLILTLA WDRVDIAKNH VFVYGQQWLV GSLEQAMLDA LVMDRVSFVK LLIENGVSMH KFLTIPRLEE LYNTK QGPT NPMLFHLIRD VKQGNLPPGY KITLIDIGLV IEYLMGGTYR CTYTRKRFRL IYNSLGGNNR RSGRNTSSST PQLRKS HET FGNRADKKEK MRHNHFIKTA QPYRPKMDAS MEEGKKKRTK DEIVDIDDPE TKRFPYPLNE LLIWACLMKR QVMARFL WQ HGEESMAKAL VACKIYRSMA YEAKQSDLVD DTSEELKQYS NDFGQLAVEL LEQSFRQDET MAMKLLTYEL KNWSNSTC L KLAVSSRLRP FVAHTCTQML LSDMWMGRLN MRKNSWYKVI LSILVPPAIL MLEYKTKAEM SHIPQSQDAH QMTMEDSEN NFHNITEEIP MEVFKEVKIL DSSDGKNEME IHIKSKKLPI TRKFYAFYHA PIVKFWFNTL AYLGFLMLYT FVVLVKMEQL PSVQEWIVI AYIFTYAIEK VREVFMSEAG KISQKIKVWF SDYFNVSDTI AIISFFVGFG LRFGAKWNYI NAYDNHVFVA G RLIYCLNI IFWYVRLLDF LAVNQQAGPY VMMIGKMVAN MFYIVVIMAL VLLSFGVPRK AILYPHEEPS WSLAKDIVFH PY WMIFGEV YAYEIDVCAN DSTLPTICGP GTWLTPFLQA VYLFVQYIIM VNLLIAFFNN VYLQVKAISN IVWKYQRYHF IMA YHEKPV LPPPLIILSH IVSLFCCVCK RRKKDKTSDG PKLFLTEEDQ KKLHDFEEQC VEMYFDEKDD KFNSGSEERI RVTF ERVEQ MSIQIKEVGD RVNYIKRSLQ SLDS UniProtKB: Transient receptor potential cation channel subfamily M member 7 |
-Macromolecule #2: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 2 / Number of copies: 12 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK I |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 BASE (4k x 4k) / Average electron dose: 56.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)