[English] 日本語
 Yorodumi
Yorodumi- EMDB-6859: Cryo-EM structure of L-fucokinase,GDP-fucose pyrophosphorylase (F... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6859 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of L-fucokinase,GDP-fucose pyrophosphorylase (FKP) in Bacteroides fragilis | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bifunctional / kinase / ATP / GTP / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Bacteroides fragilis (bacteria) Bacteroides fragilis (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Wang J / Hu H | |||||||||
 Citation Citation |  Journal: Protein Cell / Year: 2019 Journal: Protein Cell / Year: 2019Title: Cryo-EM structure of L-fucokinase/GDP-fucose pyrophosphorylase (FKP) in Bacteroides fragilis. Authors: Ying Liu / Huifang Hu / Jia Wang / Qiang Zhou / Peng Wu / Nieng Yan / Hong-Wei Wang / Jia-Wei Wu / Linfeng Sun /   | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6859.map.gz emd_6859.map.gz | 5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6859-v30.xml emd-6859-v30.xml emd-6859.xml emd-6859.xml | 9.7 KB 9.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6859_fsc.xml emd_6859_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_6859.png emd_6859.png | 88.6 KB | ||
| Filedesc metadata |  emd-6859.cif.gz emd-6859.cif.gz | 5.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6859 http://ftp.pdbj.org/pub/emdb/structures/EMD-6859 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6859 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6859 | HTTPS FTP |
-Validation report
| Summary document |  emd_6859_validation.pdf.gz emd_6859_validation.pdf.gz | 393.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6859_full_validation.pdf.gz emd_6859_full_validation.pdf.gz | 392.6 KB | Display | |
| Data in XML |  emd_6859_validation.xml.gz emd_6859_validation.xml.gz | 10.6 KB | Display | |
| Data in CIF |  emd_6859_validation.cif.gz emd_6859_validation.cif.gz | 13.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6859 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6859 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6859 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6859 | HTTPS FTP |
-Related structure data
| Related structure data |  5yysMC  6860C  6861C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6859.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6859.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
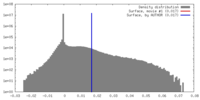
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : FKP
| Entire | Name: FKP |
|---|---|
| Components |
|
-Supramolecule #1: FKP
| Supramolecule | Name: FKP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Bacteroides fragilis (bacteria) Bacteroides fragilis (bacteria) |
-Macromolecule #1: L-fucokinase, L-fucose-1-P guanylyltransferase
| Macromolecule | Name: L-fucokinase, L-fucose-1-P guanylyltransferase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacteroides fragilis (bacteria) Bacteroides fragilis (bacteria) |
| Molecular weight | Theoretical: 105.782789 KDa |
| Sequence | String: MQKLLSLPSN LVQSFHELER VNRTDWFCTS DPVGKKLGSG GGTSWLLEEC YNEYSDGATF GEWLEKEKRI LLHAGGQSRR LPGYAPSGK ILTPVPVFRW ERGQHLGQNL LSLQLPLYEK IMSLAPDKLH TLIASGDVYI RSEKPLQSIP EADVVCYGLW V DPSLATHH ...String: MQKLLSLPSN LVQSFHELER VNRTDWFCTS DPVGKKLGSG GGTSWLLEEC YNEYSDGATF GEWLEKEKRI LLHAGGQSRR LPGYAPSGK ILTPVPVFRW ERGQHLGQNL LSLQLPLYEK IMSLAPDKLH TLIASGDVYI RSEKPLQSIP EADVVCYGLW V DPSLATHH GVFASDRKHP EQLDFMLQKP SLAELESLSK THLFLMDIGI WLLSDRAVEI LMKRSHKESS EELKYYDLYS DF GLALGTH PRIEDEEVNT LSVAILPLPG GEFYHYGTSK ELISSTLSVQ NKVYDQRRIM HRKVKPNPAM FVQNAVVRIP LCA ENADLW IENSHIGPKW KIASRHIITG VPENDWSLAV PAGVCVDVVP MGDKGFVARP YGLDDVFKGD LRDSKTTLTG IPFG EWMSK RGLSYTDLKG RTDDLQAVSV FPMVNSVEEL GLVLRWMLSE PELEEGKNIW LRSEHFSADE ISAGANLKRL YAQRE EFRK GNWKALAVNH EKSVFYQLDL ADAAEDFVRL GLDMPELLPE DALQMSRIHN RMLRARILKL DGKDYRPEEQ AAFDLL RDG LLDGISNRKS TPKLDVYSDQ IVWGRSPVRI DMAGGWTDTP PYSLYSGGNV VNLAIELNGQ PPLQVYVKPC KDFHIVL RS IDMGAMEIVS TFDELQDYKK IGSPFSIPKA ALSLAGFAPA FSAVSYASLE EQLKDFGAGI EVTLLAAIPA GSGLGTSS I LASTVLGAIN DFCGLAWDKN EICQRTLVLE QLLTTGGGWQ DQYGGVLQGV KLLQTEAGFA QSPLVRWLPD HLFTHPEYK DCHLLYYTGI TRTAKGILAE IVSSMFLNSS LHLNLLSEMK AHALDMNEAI QRGSFVEFGR LVGKTWEQNK ALDSGTNPPA VEAIIDLIK DYTLGYKLPG AGGGGYLYMV AKDPQAAVRI RKILTENAPN PRARFVEMTL SDKGFQVSRS UniProtKB: L-fucokinase/L-fucose-1-P guanylyltransferase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)