+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Endophilin B1 dimers bound to nanodiscs | |||||||||
 Map data Map data | C1 consensus map of Endophilin B1 dimers bound to a nanodisc. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | BAR / N-BAR / endophilin / membrane / curvature / cardiolipin / MSP2N2 / nanodisc / peripheral membrane protein / APOPTOSIS | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of membrane tubulation / autophagic cell death / protein localization to vacuolar membrane / positive regulation of autophagosome assembly / receptor catabolic process / membrane fission / membrane organization / positive regulation of protein targeting to mitochondrion / autophagosome membrane / regulation of macroautophagy ...positive regulation of membrane tubulation / autophagic cell death / protein localization to vacuolar membrane / positive regulation of autophagosome assembly / receptor catabolic process / membrane fission / membrane organization / positive regulation of protein targeting to mitochondrion / autophagosome membrane / regulation of macroautophagy / positive regulation of autophagy / cellular response to glucose starvation / cellular response to amino acid starvation / regulation of cytokinesis / positive regulation of protein-containing complex assembly / regulation of protein stability / autophagy / midbody / cytoplasmic vesicle / mitochondrial outer membrane / cadherin binding / Golgi membrane / lipid binding / apoptotic process / protein homodimerization activity / protein-containing complex / identical protein binding / membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
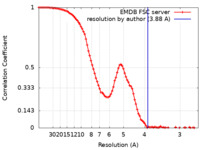
| Method | single particle reconstruction / cryo EM / Resolution: 3.88 Å | |||||||||
 Authors Authors | Thorlacius A / Sundborger-Lunna A | |||||||||
| Funding support |  Sweden, 1 items Sweden, 1 items
| |||||||||
 Citation Citation |  Journal: Biorxiv / Year: 2024 Journal: Biorxiv / Year: 2024Title: Peripheral membrane protein endophilin B1 probes, perturbs and permeabilizes lipid bilayers Authors: Thorlacius A / Rulev M / Sundberg O / Sundborger-Lunna A | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50981.map.gz emd_50981.map.gz | 31.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50981-v30.xml emd-50981-v30.xml emd-50981.xml emd-50981.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_50981_fsc.xml emd_50981_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_50981.png emd_50981.png | 70.5 KB | ||
| Masks |  emd_50981_msk_1.map emd_50981_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-50981.cif.gz emd-50981.cif.gz | 6 KB | ||
| Others |  emd_50981_half_map_1.map.gz emd_50981_half_map_1.map.gz emd_50981_half_map_2.map.gz emd_50981_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50981 http://ftp.pdbj.org/pub/emdb/structures/EMD-50981 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50981 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50981 | HTTPS FTP |
-Validation report
| Summary document |  emd_50981_validation.pdf.gz emd_50981_validation.pdf.gz | 840.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_50981_full_validation.pdf.gz emd_50981_full_validation.pdf.gz | 839.6 KB | Display | |
| Data in XML |  emd_50981_validation.xml.gz emd_50981_validation.xml.gz | 14.7 KB | Display | |
| Data in CIF |  emd_50981_validation.cif.gz emd_50981_validation.cif.gz | 20.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50981 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50981 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50981 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50981 | HTTPS FTP |
-Related structure data
| Related structure data |  9g2rMC  9g2uC  9g2wC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_50981.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50981.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C1 consensus map of Endophilin B1 dimers bound to a nanodisc. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.328 Å | ||||||||||||||||||||||||||||||||||||
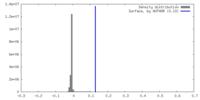
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_50981_msk_1.map emd_50981_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
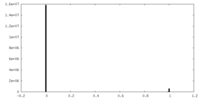
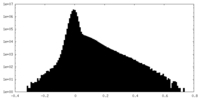
| Density Histograms |
-Half map: #2
| File | emd_50981_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
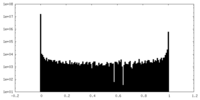
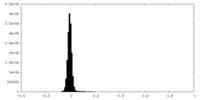
| Density Histograms |
-Half map: #1
| File | emd_50981_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
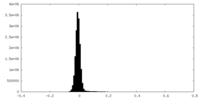
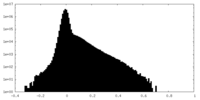
| Density Histograms |
- Sample components
Sample components
-Entire : Endophilin B1 dimers bound to a nanodisc
| Entire | Name: Endophilin B1 dimers bound to a nanodisc |
|---|---|
| Components |
|
-Supramolecule #1: Endophilin B1 dimers bound to a nanodisc
| Supramolecule | Name: Endophilin B1 dimers bound to a nanodisc / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Endophilin-B1
| Macromolecule | Name: Endophilin-B1 / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.843246 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNIMDFNVKK LAADAGTFLS RAVQFTEEKL GQAEKTELDA HLENLLSKAE CTKIWTEKIM KQTEVLLQPN PNARIEEFVY EKLDRKAPS RINNPELLGQ YMIDAGTEFG PGTAYGNALI KCGETQKRIG TADRELIQTS ALNFLTPLRN FIEGDYKTIA K ERKLLQNK ...String: MNIMDFNVKK LAADAGTFLS RAVQFTEEKL GQAEKTELDA HLENLLSKAE CTKIWTEKIM KQTEVLLQPN PNARIEEFVY EKLDRKAPS RINNPELLGQ YMIDAGTEFG PGTAYGNALI KCGETQKRIG TADRELIQTS ALNFLTPLRN FIEGDYKTIA K ERKLLQNK RLDLDAAKTR LKKAKAAETR NSSEQELRIT QSEFDRQAEI TRLLLEGISS THAHHLRCLN DFVEAQMTYY AQ CYQYMLD LQKQLGSFPS NYLSNNNQTS VTPVPSVLPN AIGSSAMAST SGLVITSPSN LSDLKECSGS RKARVLYDYD AAN STELSL LADEVITVFS VVGMDSDWLM GERGNQKGKV PITYLELLN UniProtKB: Endophilin-B1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.8 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 20 mM Tris-HCl, 100 mM NaCl, 0.5 mM EDTA, pH 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 130000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Residue range: 11-252 / Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Details | Experimental models determined from locally refined maps were rigid body fitted into the consensus map. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-9g2r: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)