+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of cardiac collagen-associated amyloid AL59 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | systemic AL amyloid fibril / PROTEIN FIBRIL | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
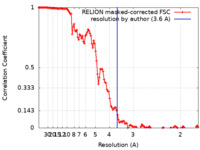
| Method | helical reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Schulte T / Speranzini V / Chaves-Sanjuan A / Milazzo M / Ricagno S | |||||||||
| Funding support |  Italy, 1 items Italy, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Helical superstructures between amyloid and collagen in cardiac fibrils from a patient with AL amyloidosis. Authors: Tim Schulte / Antonio Chaves-Sanjuan / Valentina Speranzini / Kevin Sicking / Melissa Milazzo / Giulia Mazzini / Paola Rognoni / Serena Caminito / Paolo Milani / Chiara Marabelli / ...Authors: Tim Schulte / Antonio Chaves-Sanjuan / Valentina Speranzini / Kevin Sicking / Melissa Milazzo / Giulia Mazzini / Paola Rognoni / Serena Caminito / Paolo Milani / Chiara Marabelli / Alessandro Corbelli / Luisa Diomede / Fabio Fiordaliso / Luigi Anastasia / Carlo Pappone / Giampaolo Merlini / Martino Bolognesi / Mario Nuvolone / Rubén Fernández-Busnadiego / Giovanni Palladini / Stefano Ricagno /     Abstract: Systemic light chain (LC) amyloidosis (AL) is a disease where organs are damaged by an overload of a misfolded patient-specific antibody-derived LC, secreted by an abnormal B cell clone. The high LC ...Systemic light chain (LC) amyloidosis (AL) is a disease where organs are damaged by an overload of a misfolded patient-specific antibody-derived LC, secreted by an abnormal B cell clone. The high LC concentration in the blood leads to amyloid deposition at organ sites. Indeed, cryogenic electron microscopy (cryo-EM) has revealed unique amyloid folds for heart-derived fibrils taken from different patients. Here, we present the cryo-EM structure of heart-derived AL amyloid (AL59) from another patient with severe cardiac involvement. The double-layered structure displays a u-shaped core that is closed by a β-arc lid and extended by a straight tail. Noteworthy, the fibril harbours an extended constant domain fragment, thus ruling out the variable domain as sole amyloid building block. Surprisingly, the fibrils were abundantly concatenated with a proteinaceous polymer, here identified as collagen VI (COLVI) by immuno-electron microscopy (IEM) and mass-spectrometry. Cryogenic electron tomography (cryo-ET) showed how COLVI wraps around the amyloid forming a helical superstructure, likely stabilizing and protecting the fibrils from clearance. Thus, here we report structural evidence of interactions between amyloid and collagen, potentially signifying a distinct pathophysiological mechanism of amyloid deposits. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50270.map.gz emd_50270.map.gz | 9.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50270-v30.xml emd-50270-v30.xml emd-50270.xml emd-50270.xml | 18.6 KB 18.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_50270_fsc.xml emd_50270_fsc.xml | 12.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_50270.png emd_50270.png | 103.2 KB | ||
| Masks |  emd_50270_msk_1.map emd_50270_msk_1.map | 149.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-50270.cif.gz emd-50270.cif.gz | 6.2 KB | ||
| Others |  emd_50270_half_map_1.map.gz emd_50270_half_map_1.map.gz emd_50270_half_map_2.map.gz emd_50270_half_map_2.map.gz | 118.2 MB 118.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50270 http://ftp.pdbj.org/pub/emdb/structures/EMD-50270 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50270 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50270 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_50270.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50270.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.889 Å | ||||||||||||||||||||||||||||||||||||
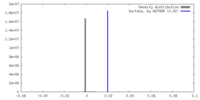
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_50270_msk_1.map emd_50270_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
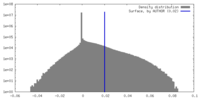
| Density Histograms |
-Half map: #1
| File | emd_50270_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
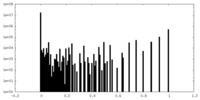
| Density Histograms |
-Half map: #2
| File | emd_50270_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
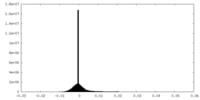
| Density Histograms |
- Sample components
Sample components
-Entire : Monoclonal immunoglobulin light chains (LC), fibrillar form
| Entire | Name: Monoclonal immunoglobulin light chains (LC), fibrillar form |
|---|---|
| Components |
|
-Supramolecule #1: Monoclonal immunoglobulin light chains (LC), fibrillar form
| Supramolecule | Name: Monoclonal immunoglobulin light chains (LC), fibrillar form type: tissue / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Amyloid fibrils extracted ex vivo from cardiac tissue of AL patient |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Monoclonal immunoglobulin light chains (LC)
| Macromolecule | Name: Monoclonal immunoglobulin light chains (LC) / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.923335 KDa |
| Sequence | String: SFELTQPSSV SVSPGQTANI TCSGGYLGET YRSWYQQKPG QSPVLVIYQS SKRPSGIPGR FSGSNSGNTA TLTISGTQPL DEADYFCQA WDFTSVVFGG GTKLTVLGQP KAAPSVTLFP PSSEELQANK ATL |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 5 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number real images: 2049 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 120000 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)