+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bufavirus 1 at pH 2.6 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Bufavirus / parvovirus / VIRUS LIKE PARTICLE | |||||||||
| Function / homology | Phospholipase A2-like domain / Phospholipase A2-like domain / Parvovirus coat protein VP2 / Parvovirus coat protein VP1/VP2 / Parvovirus coat protein VP2 / Capsid/spike protein, ssDNA virus / T=1 icosahedral viral capsid / structural molecule activity / VP1 Function and homology information Function and homology information | |||||||||
| Biological species |  Bufavirus-1 Bufavirus-1 | |||||||||
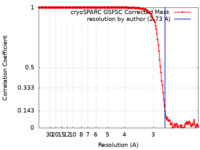
| Method | single particle reconstruction / cryo EM / Resolution: 2.73 Å | |||||||||
 Authors Authors | Gulkis MC / McKenna R / Bennett AD | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Viruses / Year: 2024 Journal: Viruses / Year: 2024Title: Structural Characterization of Human Bufavirus 1: Receptor Binding and Endosomal pH-Induced Changes. Authors: Mitchell Gulkis / Mengxiao Luo / Paul Chipman / Mario Mietzsch / Maria Söderlund-Venermo / Antonette Bennett / Robert McKenna /   Abstract: Bufaviruses (BuV) are members of the of the genus. They are non-enveloped, T = 1 icosahedral ssDNA viruses isolated from patients exhibiting acute diarrhea. The lack of treatment options and a ...Bufaviruses (BuV) are members of the of the genus. They are non-enveloped, T = 1 icosahedral ssDNA viruses isolated from patients exhibiting acute diarrhea. The lack of treatment options and a limited understanding of their disease mechanisms require studying these viruses on a molecular and structural level. In the present study, we utilize glycan arrays and cell binding assays to demonstrate that BuV1 capsid binds terminal sialic acid (SIA) glycans. Furthermore, using cryo-electron microscopy (cryo-EM), SIA is shown to bind on the 2/5-fold wall of the capsid surface. Interestingly, the capsid residues stabilizing SIA binding are conserved in all human BuVs identified to date. Additionally, biophysical assays illustrate BuV1 capsid stabilization during endo-lysosomal (pH 7.4-pH 4) trafficking and capsid destabilization at pH 3 and less, which correspond to the pH of the stomach. Hence, we determined the cryo-EM structures of BuV1 capsids at pH 7.4, 4.0, and 2.6 to 2.8 Å, 3.2 Å, and 2.7 Å, respectively. These structures reveal capsid structural rearrangements during endo-lysosomal escape and provide a potential mechanism for this process. The structural insights gained from this study will add to the general knowledge of human pathogenic parvoviruses. Furthermore, the identification of the conserved SIA receptor binding site among BuVs provides a possible targetable surface-accessible pocket for the design of small molecules to be developed as anti-virals for these viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45973.map.gz emd_45973.map.gz | 194.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45973-v30.xml emd-45973-v30.xml emd-45973.xml emd-45973.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_45973_fsc.xml emd_45973_fsc.xml | 15.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_45973.png emd_45973.png | 97.8 KB | ||
| Filedesc metadata |  emd-45973.cif.gz emd-45973.cif.gz | 6.8 KB | ||
| Others |  emd_45973_half_map_1.map.gz emd_45973_half_map_1.map.gz emd_45973_half_map_2.map.gz emd_45973_half_map_2.map.gz | 364.4 MB 364.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45973 http://ftp.pdbj.org/pub/emdb/structures/EMD-45973 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45973 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45973 | HTTPS FTP |
-Validation report
| Summary document |  emd_45973_validation.pdf.gz emd_45973_validation.pdf.gz | 870.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_45973_full_validation.pdf.gz emd_45973_full_validation.pdf.gz | 870.1 KB | Display | |
| Data in XML |  emd_45973_validation.xml.gz emd_45973_validation.xml.gz | 24.8 KB | Display | |
| Data in CIF |  emd_45973_validation.cif.gz emd_45973_validation.cif.gz | 31.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45973 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45973 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45973 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45973 | HTTPS FTP |
-Related structure data
| Related structure data |  9cwsMC  9cuzC  9cv0C  9cv9C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_45973.map.gz / Format: CCP4 / Size: 396.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45973.map.gz / Format: CCP4 / Size: 396.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.056 Å | ||||||||||||||||||||||||||||||||||||
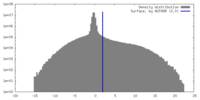
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_45973_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
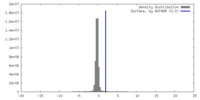
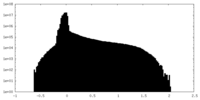
| Density Histograms |
-Half map: #1
| File | emd_45973_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
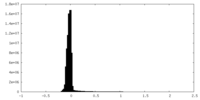
| Density Histograms |
- Sample components
Sample components
-Entire : Bufavirus-1
| Entire | Name:  Bufavirus-1 Bufavirus-1 |
|---|---|
| Components |
|
-Supramolecule #1: Bufavirus-1
| Supramolecule | Name: Bufavirus-1 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Bufavirus 1 VP2 was overexpressed in Sf9 cells / NCBI-ID: 1209382 / Sci species name: Bufavirus-1 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 4 MDa |
| Virus shell | Shell ID: 1 / Diameter: 260.0 Å / T number (triangulation number): 1 |
-Macromolecule #1: VP1
| Macromolecule | Name: VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 60 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bufavirus-1 Bufavirus-1 |
| Molecular weight | Theoretical: 80.543203 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPAIRKARGW VPPGYNYLGP FNQDFNKKPT NPSDNAARKH DLEYNKLIKQ GHNPYWNYNH ADEDFIKETD QATDWGGKFG NFVFRAKRA LAPELAPPAK KKTKTKHTEP EYSHKHIKAG TKRGKPFYLF VNLARKKARM TDTQDVSEQQ SDQPSVASTN A KAGGGGGG ...String: MPAIRKARGW VPPGYNYLGP FNQDFNKKPT NPSDNAARKH DLEYNKLIKQ GHNPYWNYNH ADEDFIKETD QATDWGGKFG NFVFRAKRA LAPELAPPAK KKTKTKHTEP EYSHKHIKAG TKRGKPFYLF VNLARKKARM TDTQDVSEQQ SDQPSVASTN A KAGGGGGG GGSGVGHSTG NYNNRTEFYY HGDEVTIVCH SSRHIHLNMS ESEEYKIYDT DRGPRFPTDQ TLQGRDTIND SY HAQVETP WFLINPNSWG TWMNPADFQQ LTTTCREVTL EHLDQTLDNI VIKTVSKQGS GAEETTQYNN DLTALLQVAL DKS NQLPWV ADNMYLDSLG YIPWRPCKLK QYSYHVNFWN TIDIISGPQQ NQWQQVKKEI RWDDLQFTPI ETTTEIDLLR TGDS WTSGP YKFNTKPTQL SYHWQSTRHT GSVHPTDPPN AIGQQGQNIR DINGWQWGDR SDPMSAATRV SNFHIGYSWP EWRIH YGSG GPAINPGAPF SQAPWSTDPQ VRLTQGASEK AIFDYNHGDD DPAHRDQWWQ NNLPITGQTN WAPKNAHQAN LSSNVP SRQ EFWTQDYHNT FGPFTAVDDV GIQYPWGAIW TKTPDTTHKP MMSAHAPFIC KDGPPGQLLV KLAPNYTENL QTDGLGN NR IVTYATFWWT GKLILKGKLR LPRQFNLYNL PGRPRGTEAK KFLPNEIGHF ELPFMPGRCM PNYTM UniProtKB: VP1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 2 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 25 sec. / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Software | Name: Leginon |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-50 / Number real images: 1496 / Average electron dose: 75.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.9 µm / Nominal defocus min: 0.1 µm / Nominal magnification: 130000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)