[English] 日本語
 Yorodumi
Yorodumi- EMDB-44479: Cryo-EM structure of synthetic claudin-4 complex with Clostridium... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of synthetic claudin-4 complex with Clostridium perfringens enterotoxin C-terminal domain, sFab COP-2, and Nanobody | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Claudin / Fab / Toxin / MEMBRANE PROTEIN / MEMBRANE PROTEIN-IMMUNE SYSYTEM complex | |||||||||
| Function / homology | Molybdenum cofactor biosynthesis protein MoaB / MoaB/Mog domain / MoaB/Mog-like domain superfamily / Probable molybdopterin binding domain / Probable molybdopterin binding domain / Mo-molybdopterin cofactor biosynthetic process / cytosol / Molybdopterin biosynthesis protein MoaB Function and homology information Function and homology information | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.16 Å | |||||||||
 Authors Authors | Vecchio AJ | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Computational design of soluble and functional membrane protein analogues. Authors: Casper A Goverde / Martin Pacesa / Nicolas Goldbach / Lars J Dornfeld / Petra E M Balbi / Sandrine Georgeon / Stéphane Rosset / Srajan Kapoor / Jagrity Choudhury / Justas Dauparas / ...Authors: Casper A Goverde / Martin Pacesa / Nicolas Goldbach / Lars J Dornfeld / Petra E M Balbi / Sandrine Georgeon / Stéphane Rosset / Srajan Kapoor / Jagrity Choudhury / Justas Dauparas / Christian Schellhaas / Simon Kozlov / David Baker / Sergey Ovchinnikov / Alex J Vecchio / Bruno E Correia /   Abstract: De novo design of complex protein folds using solely computational means remains a substantial challenge. Here we use a robust deep learning pipeline to design complex folds and soluble analogues of ...De novo design of complex protein folds using solely computational means remains a substantial challenge. Here we use a robust deep learning pipeline to design complex folds and soluble analogues of integral membrane proteins. Unique membrane topologies, such as those from G-protein-coupled receptors, are not found in the soluble proteome, and we demonstrate that their structural features can be recapitulated in solution. Biophysical analyses demonstrate the high thermal stability of the designs, and experimental structures show remarkable design accuracy. The soluble analogues were functionalized with native structural motifs, as a proof of concept for bringing membrane protein functions to the soluble proteome, potentially enabling new approaches in drug discovery. In summary, we have designed complex protein topologies and enriched them with functionalities from membrane proteins, with high experimental success rates, leading to a de facto expansion of the functional soluble fold space. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44479.map.gz emd_44479.map.gz | 114.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44479-v30.xml emd-44479-v30.xml emd-44479.xml emd-44479.xml | 23.4 KB 23.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_44479_fsc.xml emd_44479_fsc.xml | 13 KB | Display |  FSC data file FSC data file |
| Images |  emd_44479.png emd_44479.png | 52.6 KB | ||
| Filedesc metadata |  emd-44479.cif.gz emd-44479.cif.gz | 7 KB | ||
| Others |  emd_44479_half_map_1.map.gz emd_44479_half_map_1.map.gz emd_44479_half_map_2.map.gz emd_44479_half_map_2.map.gz | 213 MB 213 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44479 http://ftp.pdbj.org/pub/emdb/structures/EMD-44479 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44479 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44479 | HTTPS FTP |
-Related structure data
| Related structure data |  9beiMC  8oysC  8oyvC  8oywC  8oyxC  8oyyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_44479.map.gz / Format: CCP4 / Size: 229.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44479.map.gz / Format: CCP4 / Size: 229.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.884 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_44479_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_44479_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Synthetic human claudin-4 complex with Clostridium perfringens en...
| Entire | Name: Synthetic human claudin-4 complex with Clostridium perfringens enterotoxin C-terminal domain, sFab COP-2, and nanobody against COP-2 |
|---|---|
| Components |
|
-Supramolecule #1: Synthetic human claudin-4 complex with Clostridium perfringens en...
| Supramolecule | Name: Synthetic human claudin-4 complex with Clostridium perfringens enterotoxin C-terminal domain, sFab COP-2, and nanobody against COP-2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Assembled complex of 5 proteins (Fab is 2 proteins) expressed from insect cells and E coli |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 103 KDa |
-Macromolecule #1: Claudin-4
| Macromolecule | Name: Claudin-4 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.078986 KDa |
| Sequence | String: MSSLETFREA RRLAREGLEL VREAARLPMW RVTAFIGSNI VTSQTIWEGL WMNCVVQSTG QMQCKVYDSL LALPEDLRRA RESFERAIE VAEKALELLE IGDPDSDAIE DEEERLQTIH EAGELLLKAA ELAREPTEAI ADRIIQDFYN PLVASGQKRE M GASLALAR ...String: MSSLETFREA RRLAREGLEL VREAARLPMW RVTAFIGSNI VTSQTIWEGL WMNCVVQSTG QMQCKVYDSL LALPEDLRRA RESFERAIE VAEKALELLE IGDPDSDAIE DEEERLQTIH EAGELLLKAA ELAREPTEAI ADRIIQDFYN PLVASGQKRE M GASLALAR RGAELLEEAG RKLLGLEGGS LEHHHHHH |
-Macromolecule #2: Heat-labile enterotoxin B chain
| Macromolecule | Name: Heat-labile enterotoxin B chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.591295 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSTDIEKEIL DLAAATERLN LTDALNSNPA GNLYDWRSSN SYPWTQKLNL HLTITATGQK YRILASKIVD FNIYSNNFNN LVKLEQSLG DGVKDHYVDI SLDAGQYVLV MKANSSYSGN YPYSILFQKF UniProtKB: Molybdopterin biosynthesis protein MoaB |
-Macromolecule #3: COP-2 Fab Heavy chain
| Macromolecule | Name: COP-2 Fab Heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.26301 KDa |
| Sequence | String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NFSSSSIHWV RQAPGKGLEW VASISSYSGY TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARYWSWYNSS HYIYSALDYW GQGTLVTVSS ASTKGPSVFP LAPSSKSTSG GTAALGCLVK D YFPEPVTV ...String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NFSSSSIHWV RQAPGKGLEW VASISSYSGY TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARYWSWYNSS HYIYSALDYW GQGTLVTVSS ASTKGPSVFP LAPSSKSTSG GTAALGCLVK D YFPEPVTV SWNSGALTSG VHTFPAVLQS SGLYSLSSVV TVPSSSLGTQ TYICNVNHKP SNTKVDKKVE PKSCDKTHT |
-Macromolecule #4: Anti-fab nanobody
| Macromolecule | Name: Anti-fab nanobody / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.175438 KDa |
| Sequence | String: GSVQLQESGG GLVQPGGSLR LSCAASGRTI SRYAMSWFRQ APGKEREFVA VARRSGDGAF YADSVQGRFT VSRDDAKNTV YLQMNSLKP EDTAVYYCAI DSDTFYSGSY DYWGQGTQVT VS |
-Macromolecule #5: COP-2 Fab Light chain
| Macromolecule | Name: COP-2 Fab Light chain / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.517057 KDa |
| Sequence | String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQSYEWAPVT FGQGTKVEIK RTVAAPSVFI FPPSDSQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ ...String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQSYEWAPVT FGQGTKVEIK RTVAAPSVFI FPPSDSQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ DSKDSTYSLS STLTLSKADY EKHKVYACEV THQGLSSPVT KSFNRGEC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: 20 mM Hepes pH 8.0, 150 mM NaCl | |||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER Details: UltraAuFoil 1.2/1.3 grids (Quantifoil) were glow discharged for 30 s at 15 mA in a Pelco easiGlow (Ted Pella Inc) instrument | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: LEICA EM GP Details: 3.5 microL of complex was applied onto grids and blotted for 3 s at 4 degrees C under 100 percent humidity then plunge frozen into liquid ethane cooled by liquid nitrogen.. |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 1159 / Average electron dose: 49.4 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.4 µm / Nominal magnification: 120000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 322 | |||||||||||||||
| Output model |  PDB-9bei: |
 Movie
Movie Controller
Controller


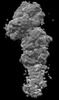
 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)