登録情報 データベース : EMDB / ID : EMD-43196タイトル Cryo-EM structure of GATA4 in complex with ALBN1 nucleosome main map 複合体 : GATA4 in complex with 186bp ALBN1 nucleosomeDNA : DNA (159-MER)DNA : DNA (159-MER)タンパク質・ペプチド : Histone H3.1タンパク質・ペプチド : Histone H4タンパク質・ペプチド : Histone H2A type 1-B/Eタンパク質・ペプチド : Histone H2B type 1-Jタンパク質・ペプチド : Maltose/maltodextrin-binding periplasmic protein,Transcription factor GATA-4リガンド : ZINC ION / / / / / / 機能・相同性 分子機能 ドメイン・相同性 構成要素
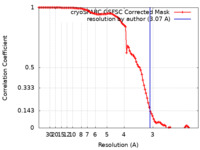
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Homo sapiens (ヒト) / synthetic construct (人工物) 手法 / / 解像度 : 3.07 Å Zhou BR / Bai Y 資金援助 Organization Grant number 国 National Institutes of Health/National Cancer Institute (NIH/NCI) Intramural Research Program
ジャーナル : Mol Cell / 年 : 2024タイトル : Structural insights into the cooperative nucleosome recognition and chromatin opening by FOXA1 and GATA4.著者 : Bing-Rui Zhou / Hanqiao Feng / Furong Huang / Iris Zhu / Stephanie Portillo-Ledesma / Dan Shi / Kenneth S Zaret / Tamar Schlick / David Landsman / Qianben Wang / Yawen Bai / 要旨 : Mouse FOXA1 and GATA4 are prototypes of pioneer factors, initiating liver cell development by binding to the N1 nucleosome in the enhancer of the ALB1 gene. Using cryoelectron microscopy (cryo-EM), ... Mouse FOXA1 and GATA4 are prototypes of pioneer factors, initiating liver cell development by binding to the N1 nucleosome in the enhancer of the ALB1 gene. Using cryoelectron microscopy (cryo-EM), we determined the structures of the free N1 nucleosome and its complexes with FOXA1 and GATA4, both individually and in combination. We found that the DNA-binding domains of FOXA1 and GATA4 mainly recognize the linker DNA and an internal site in the nucleosome, respectively, whereas their intrinsically disordered regions interact with the acidic patch on histone H2A-H2B. FOXA1 efficiently enhances GATA4 binding by repositioning the N1 nucleosome. In vivo DNA editing and bioinformatics analyses suggest that the co-binding mode of FOXA1 and GATA4 plays important roles in regulating genes involved in liver cell functions. Our results reveal the mechanism whereby FOXA1 and GATA4 cooperatively bind to the nucleosome through nucleosome repositioning, opening chromatin by bending linker DNA and obstructing nucleosome packing. 履歴 登録 2023年12月22日 - ヘッダ(付随情報) 公開 2024年8月7日 - マップ公開 2024年8月7日 - 更新 2024年9月11日 - 現状 2024年9月11日 処理サイト : RCSB / 状態 : 公開
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報
 マップデータ
マップデータ 試料
試料 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Homo sapiens (ヒト) / synthetic construct (人工物)
Homo sapiens (ヒト) / synthetic construct (人工物) データ登録者
データ登録者 米国, 1件
米国, 1件  引用
引用 ジャーナル: Mol Cell / 年: 2024
ジャーナル: Mol Cell / 年: 2024

 構造の表示
構造の表示 ダウンロードとリンク
ダウンロードとリンク emd_43196.map.gz
emd_43196.map.gz EMDBマップデータ形式
EMDBマップデータ形式 emd-43196-v30.xml
emd-43196-v30.xml emd-43196.xml
emd-43196.xml EMDBヘッダ
EMDBヘッダ emd_43196_fsc.xml
emd_43196_fsc.xml FSCデータファイル
FSCデータファイル emd_43196.png
emd_43196.png emd-43196.cif.gz
emd-43196.cif.gz emd_43196_half_map_1.map.gz
emd_43196_half_map_1.map.gz emd_43196_half_map_2.map.gz
emd_43196_half_map_2.map.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-43196
http://ftp.pdbj.org/pub/emdb/structures/EMD-43196 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43196
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43196 emd_43196_validation.pdf.gz
emd_43196_validation.pdf.gz EMDB検証レポート
EMDB検証レポート emd_43196_full_validation.pdf.gz
emd_43196_full_validation.pdf.gz emd_43196_validation.xml.gz
emd_43196_validation.xml.gz emd_43196_validation.cif.gz
emd_43196_validation.cif.gz https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43196
https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43196 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43196
ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43196 リンク
リンク EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource マップ
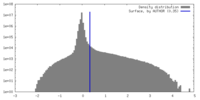
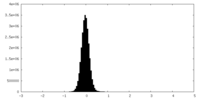
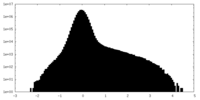
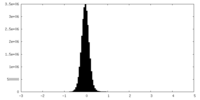
マップ ダウンロード / ファイル: emd_43196.map.gz / 形式: CCP4 / 大きさ: 103 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
ダウンロード / ファイル: emd_43196.map.gz / 形式: CCP4 / 大きさ: 103 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) 試料の構成要素
試料の構成要素 Homo sapiens (ヒト)
Homo sapiens (ヒト) Homo sapiens (ヒト)
Homo sapiens (ヒト)
 Homo sapiens (ヒト)
Homo sapiens (ヒト)
 Homo sapiens (ヒト)
Homo sapiens (ヒト)
 Homo sapiens (ヒト)
Homo sapiens (ヒト)
 Homo sapiens (ヒト)
Homo sapiens (ヒト)
 解析
解析 試料調製
試料調製 電子顕微鏡法
電子顕微鏡法 FIELD EMISSION GUN
FIELD EMISSION GUN
 ムービー
ムービー コントローラー
コントローラー


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)