+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Active conformation of DNA polymerase gamma bound to DNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA BINDING PROTEIN / DNA polymerase / mitochondrial DNA replication / DNA polymerase gamma / DNA BINDING PROTEIN-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationgamma DNA polymerase complex / mitochondrial chromosome / Strand-asynchronous mitochondrial DNA replication / mitochondrial DNA replication / positive regulation of DNA-directed DNA polymerase activity / DNA replication proofreading / single-stranded DNA 3'-5' DNA exonuclease activity / Hydrolases; Acting on ester bonds; Exodeoxyribonucleases producing 5'-phosphomonoesters / DNA metabolic process / DNA polymerase processivity factor activity ...gamma DNA polymerase complex / mitochondrial chromosome / Strand-asynchronous mitochondrial DNA replication / mitochondrial DNA replication / positive regulation of DNA-directed DNA polymerase activity / DNA replication proofreading / single-stranded DNA 3'-5' DNA exonuclease activity / Hydrolases; Acting on ester bonds; Exodeoxyribonucleases producing 5'-phosphomonoesters / DNA metabolic process / DNA polymerase processivity factor activity / mitochondrial nucleoid / Lyases; Carbon-oxygen lyases; Other carbon-oxygen lyases / 5'-deoxyribose-5-phosphate lyase activity / base-excision repair, gap-filling / DNA polymerase binding / 3'-5' exonuclease activity / Transcriptional activation of mitochondrial biogenesis / base-excision repair / DNA-templated DNA replication / protease binding / double-stranded DNA binding / in utero embryonic development / DNA-directed DNA polymerase / DNA-directed DNA polymerase activity / mitochondrial matrix / intracellular membrane-bounded organelle / chromatin binding / protein-containing complex / mitochondrion / DNA binding / identical protein binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
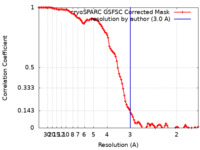
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Riccio AA / Krahn JM / Bouvette J / Borgnia JM / Copeland WC | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2024 Journal: Nucleic Acids Res / Year: 2024Title: Coordinated DNA polymerization by Polγ and the region of LonP1 regulated proteolysis. Authors: Amanda A Riccio / Asia J Brannon / Juno M Krahn / Jonathan Bouvette / Jason G Williams / Mario J Borgnia / William C Copeland /  Abstract: The replicative mitochondrial DNA polymerase, Polγ, and its protein regulation are essential for the integrity of the mitochondrial genome. The intricacies of Polγ regulation and its interactions ...The replicative mitochondrial DNA polymerase, Polγ, and its protein regulation are essential for the integrity of the mitochondrial genome. The intricacies of Polγ regulation and its interactions with regulatory proteins, which are essential for fine-tuning polymerase function, remain poorly understood. Misregulation of the Polγ heterotrimer, consisting of (i) PolG, the polymerase catalytic subunit and (ii) PolG2, the accessory subunit, ultimately results in mitochondrial diseases. Here, we used single particle cryo-electron microscopy to resolve the structure of PolG in its apoprotein state and we captured Polγ at three intermediates within the catalytic cycle: DNA bound, engaged, and an active polymerization state. Chemical crosslinking mass spectrometry, and site-directed mutagenesis uncovered the region of LonP1 engagement of PolG, which promoted proteolysis and regulation of PolG protein levels. PolG2 clinical variants, which disrupted a stable Polγ complex, led to enhanced LonP1-mediated PolG degradation. Overall, this insight into Polγ aids in an understanding of mitochondrial DNA replication and characterizes how machinery of the replication fork may be targeted for proteolytic degradation when improperly functioning. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42984.map.gz emd_42984.map.gz | 32.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42984-v30.xml emd-42984-v30.xml emd-42984.xml emd-42984.xml | 21.7 KB 21.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42984_fsc.xml emd_42984_fsc.xml | 10 KB | Display |  FSC data file FSC data file |
| Images |  emd_42984.png emd_42984.png | 54.5 KB | ||
| Filedesc metadata |  emd-42984.cif.gz emd-42984.cif.gz | 7.7 KB | ||
| Others |  emd_42984_half_map_1.map.gz emd_42984_half_map_1.map.gz emd_42984_half_map_2.map.gz emd_42984_half_map_2.map.gz | 99.2 MB 99.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42984 http://ftp.pdbj.org/pub/emdb/structures/EMD-42984 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42984 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42984 | HTTPS FTP |
-Related structure data
| Related structure data |  8v5rMC  8v54C  8v55C  8v5dC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42984.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42984.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.833 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_42984_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
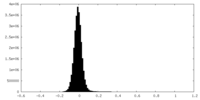
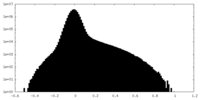
| Density Histograms |
-Half map: #1
| File | emd_42984_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
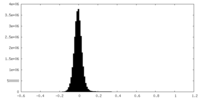
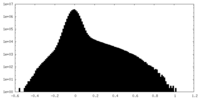
| Density Histograms |
- Sample components
Sample components
-Entire : DNA polymerase gamma complex in an active conformation
| Entire | Name: DNA polymerase gamma complex in an active conformation |
|---|---|
| Components |
|
-Supramolecule #1: DNA polymerase gamma complex in an active conformation
| Supramolecule | Name: DNA polymerase gamma complex in an active conformation type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 504 KDa |
-Macromolecule #1: DNA polymerase subunit gamma-1
| Macromolecule | Name: DNA polymerase subunit gamma-1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 138.723438 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGGSHHHHHH GSRFMVSSSV PASDPSDGQR RRQQQQQQQQ QQQQQPQQPQ VLSSEGGQLR HNPLDIQMLS RGLHEQIFGQ GGEMPGEAA VRRSVEHLQK HGLWGQPAVP LPDVELRLPP LYGDNLDQHF RLLAQKQSLP YLEAANLLLQ AQLPPKPPAW A WAEGWTRY ...String: MGGSHHHHHH GSRFMVSSSV PASDPSDGQR RRQQQQQQQQ QQQQQPQQPQ VLSSEGGQLR HNPLDIQMLS RGLHEQIFGQ GGEMPGEAA VRRSVEHLQK HGLWGQPAVP LPDVELRLPP LYGDNLDQHF RLLAQKQSLP YLEAANLLLQ AQLPPKPPAW A WAEGWTRY GPEGEAVPVA IPEERALVFA VAVCLAEGTC PTLAVAISPS AWYSWCSQRL VEERYSWTSQ LSPADLIPLE VP TGASSPT QRDWQEQLVV GHNVSFDRAH IREQYLIQGS RMRFLDTMSM HMAISGLSSF QRSLWIAAKQ GKHKVQPPTK QGQ KSQRKA RRGPAISSWD WLDISSVNSL AEVHRLYVGG PPLEKEPREL FVKGTMKDIR ENFQDLMQYC AQDVWATHEV FQQQ LPLFL ERCPHPVTLA GMLEMGVSYL PVNQNWERYL AEAQGTYEEL QREMKKSLMD LANDACQLLS GERYKEDPWL WDLEW DLQE FKQKKAKKVK KEPATASKLP IEGAGAPGDP MDQEDLGPCS EEEEFQQDVM ARACLQKLKG TTELLPKRPQ HLPGHP GWY RKLCPRLDDP AWTPGPSLLS LQMRVTPKLM ALTWDGFPLH YSERHGWGYL VPGRRDNLAK LPTGTTLESA GVVCPYR AI ESLYRKHCLE QGKQQLMPQE AGLAEEFLLT DNSAIWQTVE ELDYLEVEAE AKMENLRAAV PGQPLALTAR GGPKDTQP S YHHGNGPYND VDIPGCWFFK LPHKDGNSCN VGSPFAKDFL PKMEDGTLQA GPGGASGPRA LEINKMISFW RNAHKRISS QMVVWLPRSA LPRAVIRHPD YDEEGLYGAI LPQVVTAGTI TRRAVEPTWL TASNARPDRV GSELKAMVQA PPGYTLVGAD VDSQELWIA AVLGDAHFAG MHGCTAFGWM TLQGRKSRGT DLHSKTATTV GISREHAKIF NYGRIYGAGQ PFAERLLMQF N HRLTQQEA AEKAQQMYAA TKGLRWYRLS DEGEWLVREL NLPVDRTEGG WISLQDLRKV QRETARKSQW KKWEVVAERA WK GGTESEM FNKLESIATS DIPRTPVLGC CISRALEPSA VQEEFMTSRV NWVVQSSAVD YLHLMLVAMK WLFEEFAIDG RFC ISIHDE VRYLVREEDR YRAALALQIT NLLTRCMFAY KLGLNDLPQS VAFFSAVDID RCLRKEVTMD CKTPSNPTGM ERRY GIPQG EALDIYQIIE LTKGSLEKRS QPGP UniProtKB: DNA polymerase subunit gamma-1 |
-Macromolecule #2: DNA polymerase subunit gamma-2, mitochondrial
| Macromolecule | Name: DNA polymerase subunit gamma-2, mitochondrial / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 53.787281 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASRGSHHHH HHGADAGQPE LLTERSSPKG GHVKSHAELE GNGEHPEAPG SGEGSEALLE ICQRRHFLSG SKQQLSRDSL LSGCHPGFG PLGVELRKNL AAEWWTSVVV FREQVFPVDA LHHKPGPLLP GDSAFRLVSA ETLREILQDK ELSKEQLVAF L ENVLKTSG ...String: MASRGSHHHH HHGADAGQPE LLTERSSPKG GHVKSHAELE GNGEHPEAPG SGEGSEALLE ICQRRHFLSG SKQQLSRDSL LSGCHPGFG PLGVELRKNL AAEWWTSVVV FREQVFPVDA LHHKPGPLLP GDSAFRLVSA ETLREILQDK ELSKEQLVAF L ENVLKTSG KLRENLLHGA LEHYVNCLDL VNKRLPYGLA QIGVCFHPVF DTKQIRNGVK SIGEKTEASL VWFTPPRTSN QW LDFWLRH RLQWWRKFAM SPSNFSSSDC QDEEGRKGNK LYYNFPWGKE LIETLWNLGD HELLHMYPGN VSKLHGRDGR KNV VPCVLS VNGDLDRGML AYLYDSFQLT ENSFTRKKNL HRKVLKLHPC LAPIKVALDV GRGPTLELRQ VCQGLFNELL ENGI SVWPG YLETMQSSLE QLYSKYDEMS ILFTVLVTET TLENGLIHLR SRDTTMKEMM HISKLKDFLI KYISSAKNV UniProtKB: DNA polymerase subunit gamma-2 |
-Macromolecule #3: primer DNA
| Macromolecule | Name: primer DNA / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 15.780146 KDa |
| Sequence | String: (DT)(DG)(DG)(DG)(DT)(DT)(DT)(DT)(DT)(DA) (DT)(DG)(DT)(DA)(DC)(DT)(DA)(DC)(DA)(DG) (DG)(DT)(DG)(DG)(DT)(DC)(DA)(DA)(DG) (DT)(DA)(DT)(DT)(DT)(DA)(DT)(DG)(DG)(DT) (DA) (DC)(DC)(DG)(DT)(DA)(DC)(DA)(DA) (DT)(DA)(2DT) |
-Macromolecule #4: Template DNA
| Macromolecule | Name: Template DNA / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.477756 KDa |
| Sequence | String: (DA)(DT)(DT)(DT)(DC)(DG)(DT)(DA)(DC)(DA) (DT)(DT)(DA)(DC)(DT)(DG)(DC)(DC)(DA)(DG) (DC)(DC)(DA)(DC)(DC)(DA)(DT)(DG)(DA) (DA)(DT)(DA)(DT)(DT)(DG)(DT)(DA)(DC)(DG) (DG) (DT)(DA)(DC)(DC)(DA)(DT) ...String: (DA)(DT)(DT)(DT)(DC)(DG)(DT)(DA)(DC)(DA) (DT)(DT)(DA)(DC)(DT)(DG)(DC)(DC)(DA)(DG) (DC)(DC)(DA)(DC)(DC)(DA)(DT)(DG)(DA) (DA)(DT)(DA)(DT)(DT)(DG)(DT)(DA)(DC)(DG) (DG) (DT)(DA)(DC)(DC)(DA)(DT)(DA)(DA) (DA)(DT)(DA)(DC)(DT)(DT)(DG)(DA)(DC)(DC) (DA)(DC) (DC)(DT)(DG)(DT)(DA)(DG)(DT) (DA)(DC)(DA)(DT)(DA)(DA)(DA)(DA)(DA)(DC) (DC)(DC)(DA) |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE
| Macromolecule | Name: 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE / type: ligand / ID: 6 / Number of copies: 1 / Formula: D3T |
|---|---|
| Molecular weight | Theoretical: 466.169 Da |
| Chemical component information | 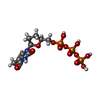 ChemComp-23T: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)