+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH in loaded state, mRNA with a 24 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | |||||||||
 マップデータ マップデータ | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH in loaded state, mRNA with a 24 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | transcription / translation / RfaH / gene expression / regulation / ops / ribosome / coupling | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報transcription antitermination factor activity, DNA binding / negative regulation of cytoplasmic translational initiation / RNA polymerase complex / transcription elongation-coupled chromatin remodeling / submerged biofilm formation / stringent response / cellular response to cell envelope stress / cytosolic DNA-directed RNA polymerase complex / bacterial-type RNA polymerase core enzyme binding / regulation of DNA-templated transcription initiation ...transcription antitermination factor activity, DNA binding / negative regulation of cytoplasmic translational initiation / RNA polymerase complex / transcription elongation-coupled chromatin remodeling / submerged biofilm formation / stringent response / cellular response to cell envelope stress / cytosolic DNA-directed RNA polymerase complex / bacterial-type RNA polymerase core enzyme binding / regulation of DNA-templated transcription initiation / bacterial-type flagellum assembly / misfolded RNA binding / Group I intron splicing / RNA folding / bacterial-type flagellum-dependent cell motility / positive regulation of ribosome biogenesis / nitrate assimilation / translational termination / DnaA-L2 complex / negative regulation of translational initiation / negative regulation of DNA-templated DNA replication initiation / mRNA regulatory element binding translation repressor activity / assembly of large subunit precursor of preribosome / positive regulation of RNA splicing / ribosome assembly / transcription elongation factor complex / cytosolic ribosome assembly / regulation of DNA-templated transcription elongation / transcription antitermination / translational initiation / DNA-templated transcription initiation / cell motility / regulation of cell growth / DNA-templated transcription termination / maintenance of translational fidelity / ribonucleoside binding / mRNA 5'-UTR binding / : / : / : / : / : / : / DNA-directed RNA polymerase / large ribosomal subunit / ribosome binding / transferase activity / ribosomal small subunit assembly / ribosomal small subunit biogenesis / response to heat / small ribosomal subunit / small ribosomal subunit rRNA binding / 5S rRNA binding / ribosomal large subunit assembly / cytosolic small ribosomal subunit / protein-containing complex assembly / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / intracellular iron ion homeostasis / cytoplasmic translation / tRNA binding / protein dimerization activity / negative regulation of translation / rRNA binding / ribosome / structural constituent of ribosome / translation / ribonucleoprotein complex / response to antibiotic / mRNA binding / magnesium ion binding / DNA binding / RNA binding / zinc ion binding / membrane / cytosol / cytoplasm 類似検索 - 分子機能 | |||||||||
| 生物種 |  | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.2 Å | |||||||||
 データ登録者 データ登録者 | Molodtsov V / Wang C / Ebright RH | |||||||||
| 資金援助 |  米国, 1件 米国, 1件
| |||||||||
 引用 引用 |  ジャーナル: Nat Struct Mol Biol / 年: 2024 ジャーナル: Nat Struct Mol Biol / 年: 2024タイトル: Structural basis of RfaH-mediated transcription-translation coupling. 著者: Vadim Molodtsov / Chengyuan Wang / Jing Zhang / Jason T Kaelber / Gregor Blaha / Richard H Ebright /    要旨: The NusG paralog RfaH mediates bacterial transcription-translation coupling in genes that contain a DNA sequence element, termed an ops site, required for pausing RNA polymerase (RNAP) and for ...The NusG paralog RfaH mediates bacterial transcription-translation coupling in genes that contain a DNA sequence element, termed an ops site, required for pausing RNA polymerase (RNAP) and for loading RfaH onto the paused RNAP. Here, we report cryo-electron microscopy structures of transcription-translation complexes (TTCs) containing Escherichia coli RfaH. The results show that RfaH bridges RNAP and the ribosome, with the RfaH N-terminal domain interacting with RNAP and the RfaH C-terminal domain interacting with the ribosome. The results show that the distribution of translational and orientational positions of RNAP relative to the ribosome in RfaH-coupled TTCs is more restricted than in NusG-coupled TTCs because of the more restricted flexibility of the RfaH interdomain linker. The results further suggest that the structural organization of RfaH-coupled TTCs in the 'loading state', in which RNAP and RfaH are located at the ops site during formation of the TTC, is the same as the structural organization of RfaH-coupled TTCs in the 'loaded state', in which RNAP and RfaH are located at positions downstream of the ops site during function of the TTC. The results define the structural organization of RfaH-containing TTCs and set the stage for analysis of functions of RfaH during translation initiation and transcription-translation coupling. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_42473.map.gz emd_42473.map.gz | 591.2 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-42473-v30.xml emd-42473-v30.xml emd-42473.xml emd-42473.xml | 93.1 KB 93.1 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_42473.png emd_42473.png | 40.3 KB | ||
| Filedesc metadata |  emd-42473.cif.gz emd-42473.cif.gz | 19.6 KB | ||
| その他 |  emd_42473_half_map_1.map.gz emd_42473_half_map_1.map.gz emd_42473_half_map_2.map.gz emd_42473_half_map_2.map.gz | 591.8 MB 591.8 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42473 http://ftp.pdbj.org/pub/emdb/structures/EMD-42473 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42473 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42473 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_42473_validation.pdf.gz emd_42473_validation.pdf.gz | 1.2 MB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_42473_full_validation.pdf.gz emd_42473_full_validation.pdf.gz | 1.2 MB | 表示 | |
| XML形式データ |  emd_42473_validation.xml.gz emd_42473_validation.xml.gz | 20.3 KB | 表示 | |
| CIF形式データ |  emd_42473_validation.cif.gz emd_42473_validation.cif.gz | 24.3 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42473 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42473 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42473 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42473 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8uqlMC  8upoC  8uprC  8uqmC  8uqpC  8ur0C  8urhC  8uriC  8urxC  8uryC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_42473.map.gz / 形式: CCP4 / 大きさ: 729 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_42473.map.gz / 形式: CCP4 / 大きさ: 729 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH in loaded state, mRNA with a 24 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.7983 Å | ||||||||||||||||||||||||||||||||||||
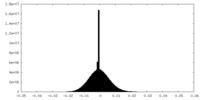
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-ハーフマップ: half-map 2
| ファイル | emd_42473_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | half-map 2 | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
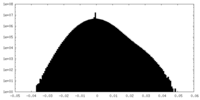
| 密度ヒストグラム |
-ハーフマップ: half-map 1
| ファイル | emd_42473_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | half-map 1 | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
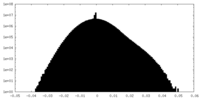
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
+全体 : Escherichia coli transcription-translation coupled complex class ...
+超分子 #1: Escherichia coli transcription-translation coupled complex class ...
+分子 #1: Ribosomal protein L21
+分子 #2: 50S ribosomal protein L22
+分子 #3: 50S ribosomal protein L23
+分子 #4: 50S ribosomal protein L24
+分子 #5: 50S ribosomal protein L25
+分子 #9: 50S ribosomal protein L10
+分子 #11: DNA-directed RNA polymerase subunit beta
+分子 #12: Transcription antitermination protein RfaH
+分子 #13: DNA-directed RNA polymerase subunit alpha
+分子 #14: DNA-directed RNA polymerase subunit beta'
+分子 #15: DNA-directed RNA polymerase subunit omega
+分子 #16: 30S ribosomal protein S18
+分子 #18: 30S ribosomal protein S20
+分子 #19: 30S ribosomal protein S21
+分子 #20: 30S ribosomal protein S2
+分子 #21: 30S ribosomal protein S1
+分子 #22: 30S ribosomal protein S3
+分子 #23: 30S ribosomal protein S4
+分子 #24: 30S ribosomal protein S5
+分子 #25: 30S ribosomal protein S6
+分子 #26: 30S ribosomal protein S7
+分子 #27: 30S ribosomal protein S8
+分子 #28: 30S ribosomal protein S9
+分子 #29: 30S ribosomal protein S10
+分子 #30: 30S ribosomal protein S11
+分子 #31: 30S ribosomal protein S12
+分子 #32: 30S ribosomal protein S14
+分子 #33: 30S ribosomal protein S15
+分子 #34: 30S ribosomal protein S16
+分子 #35: 30S ribosomal protein S17
+分子 #36: 30S ribosomal protein S19
+分子 #37: 30S ribosomal protein S13
+分子 #38: 50S ribosomal protein L11
+分子 #39: 50S ribosomal protein L7/L12
+分子 #41: 50S ribosomal protein L27
+分子 #42: 50S ribosomal protein L28
+分子 #44: 50S ribosomal protein L29
+分子 #45: 50S ribosomal protein L30
+分子 #46: 50S ribosomal protein L31
+分子 #47: 50S ribosomal protein L2
+分子 #48: 50S ribosomal protein L32
+分子 #49: 50S ribosomal protein L3
+分子 #50: 50S ribosomal protein L33
+分子 #51: 50S ribosomal protein L4
+分子 #52: 50S ribosomal protein L34
+分子 #53: 50S ribosomal protein L5
+分子 #54: 50S ribosomal protein L35
+分子 #55: 50S ribosomal protein L6
+分子 #56: 50S ribosomal protein L36
+分子 #57: 50S ribosomal protein L9
+分子 #58: 50S ribosomal protein L13
+分子 #59: 50S ribosomal protein L14
+分子 #60: 50S ribosomal protein L15
+分子 #61: 50S ribosomal protein L16
+分子 #62: 50S ribosomal protein L17
+分子 #63: 50S ribosomal protein L18
+分子 #64: 50S ribosomal protein L19
+分子 #65: 50S ribosomal protein L20
+分子 #6: NT DNA
+分子 #7: T DNA
+分子 #8: mRNA with 24 nt long spacer
+分子 #10: E-site and P-site fMet-tRNA
+分子 #17: 16S rRNA
+分子 #40: 23S rRNA
+分子 #43: 5S rRNA
+分子 #66: MAGNESIUM ION
+分子 #67: ZINC ION
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.5 |
|---|---|
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TALOS ARCTICA |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 平均電子線量: 28.0 e/Å2 |
| 電子線 | 加速電圧: 200 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 2.5 µm / 最小 デフォーカス(公称値): 1.25 µm |
| 実験機器 |  モデル: Talos Arctica / 画像提供: FEI Company |
- 画像解析
画像解析
| 初期モデル | モデルのタイプ: NONE |
|---|---|
| 最終 再構成 | 解像度のタイプ: BY AUTHOR / 解像度: 3.2 Å / 解像度の算出法: FSC 0.143 CUT-OFF / 使用した粒子像数: 35602 |
| 初期 角度割当 | タイプ: COMMON LINE |
| 最終 角度割当 | タイプ: COMMON LINE |
 ムービー
ムービー コントローラー
コントローラー























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)