[English] 日本語
 Yorodumi
Yorodumi- EMDB-42473: Escherichia coli transcription-translation coupled complex class ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH in loaded state, mRNA with a 24 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | |||||||||
 Map data Map data | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH in loaded state, mRNA with a 24 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transcription / translation / RfaH / gene expression / regulation / ops / ribosome / coupling | |||||||||
| Function / homology |  Function and homology information Function and homology informationtranscription antitermination factor activity, DNA binding / DNA-templated transcription elongation / RNA polymerase complex / negative regulation of cytoplasmic translational initiation / stringent response / submerged biofilm formation / cellular response to cell envelope stress / regulation of DNA-templated transcription initiation / bacterial-type flagellum assembly / bacterial-type RNA polymerase core enzyme binding ...transcription antitermination factor activity, DNA binding / DNA-templated transcription elongation / RNA polymerase complex / negative regulation of cytoplasmic translational initiation / stringent response / submerged biofilm formation / cellular response to cell envelope stress / regulation of DNA-templated transcription initiation / bacterial-type flagellum assembly / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / misfolded RNA binding / Group I intron splicing / RNA folding / bacterial-type flagellum-dependent cell motility / nitrate assimilation / positive regulation of ribosome biogenesis / translational termination / DnaA-L2 complex / negative regulation of translational initiation / negative regulation of DNA-templated DNA replication initiation / mRNA regulatory element binding translation repressor activity / positive regulation of RNA splicing / assembly of large subunit precursor of preribosome / cytosolic ribosome assembly / regulation of DNA-templated transcription elongation / ribosome assembly / transcription elongation factor complex / transcription antitermination / DNA-directed RNA polymerase complex / regulation of cell growth / cell motility / DNA-templated transcription initiation / translational initiation / DNA-templated transcription termination / maintenance of translational fidelity / mRNA 5'-UTR binding / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / large ribosomal subunit / transferase activity / ribosome binding / response to heat / ribosomal small subunit biogenesis / ribosomal small subunit assembly / ribosomal large subunit assembly / protein-containing complex assembly / 5S rRNA binding / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / intracellular iron ion homeostasis / cytoplasmic translation / tRNA binding / protein dimerization activity / negative regulation of translation / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / response to antibiotic / mRNA binding / DNA-templated transcription / magnesium ion binding / DNA binding / RNA binding / zinc ion binding / membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Molodtsov V / Wang C / Ebright RH | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structural basis of RfaH-mediated transcription-translation coupling. Authors: Vadim Molodtsov / Chengyuan Wang / Jing Zhang / Jason T Kaelber / Gregor Blaha / Richard H Ebright /    Abstract: The NusG paralog RfaH mediates bacterial transcription-translation coupling in genes that contain a DNA sequence element, termed an ops site, required for pausing RNA polymerase (RNAP) and for ...The NusG paralog RfaH mediates bacterial transcription-translation coupling in genes that contain a DNA sequence element, termed an ops site, required for pausing RNA polymerase (RNAP) and for loading RfaH onto the paused RNAP. Here, we report cryo-electron microscopy structures of transcription-translation complexes (TTCs) containing Escherichia coli RfaH. The results show that RfaH bridges RNAP and the ribosome, with the RfaH N-terminal domain interacting with RNAP and the RfaH C-terminal domain interacting with the ribosome. The results show that the distribution of translational and orientational positions of RNAP relative to the ribosome in RfaH-coupled TTCs is more restricted than in NusG-coupled TTCs because of the more restricted flexibility of the RfaH interdomain linker. The results further suggest that the structural organization of RfaH-coupled TTCs in the 'loading state', in which RNAP and RfaH are located at the ops site during formation of the TTC, is the same as the structural organization of RfaH-coupled TTCs in the 'loaded state', in which RNAP and RfaH are located at positions downstream of the ops site during function of the TTC. The results define the structural organization of RfaH-containing TTCs and set the stage for analysis of functions of RfaH during translation initiation and transcription-translation coupling. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42473.map.gz emd_42473.map.gz | 591.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42473-v30.xml emd-42473-v30.xml emd-42473.xml emd-42473.xml | 93.1 KB 93.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_42473.png emd_42473.png | 40.3 KB | ||
| Filedesc metadata |  emd-42473.cif.gz emd-42473.cif.gz | 19.6 KB | ||
| Others |  emd_42473_half_map_1.map.gz emd_42473_half_map_1.map.gz emd_42473_half_map_2.map.gz emd_42473_half_map_2.map.gz | 591.8 MB 591.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42473 http://ftp.pdbj.org/pub/emdb/structures/EMD-42473 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42473 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42473 | HTTPS FTP |
-Related structure data
| Related structure data |  8uqlMC  8upoC  8uprC  8uqmC  8uqpC  8ur0C  8urhC  8uriC  8urxC  8uryC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42473.map.gz / Format: CCP4 / Size: 729 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42473.map.gz / Format: CCP4 / Size: 729 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH in loaded state, mRNA with a 24 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.7983 Å | ||||||||||||||||||||||||||||||||||||
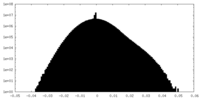
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half-map 2
| File | emd_42473_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
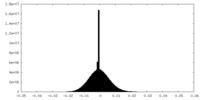
| Density Histograms |
-Half map: half-map 1
| File | emd_42473_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
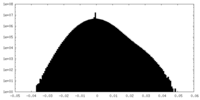
| Density Histograms |
- Sample components
Sample components
+Entire : Escherichia coli transcription-translation coupled complex class ...
+Supramolecule #1: Escherichia coli transcription-translation coupled complex class ...
+Macromolecule #1: Ribosomal protein L21
+Macromolecule #2: 50S ribosomal protein L22
+Macromolecule #3: 50S ribosomal protein L23
+Macromolecule #4: 50S ribosomal protein L24
+Macromolecule #5: 50S ribosomal protein L25
+Macromolecule #9: 50S ribosomal protein L10
+Macromolecule #11: DNA-directed RNA polymerase subunit beta
+Macromolecule #12: Transcription antitermination protein RfaH
+Macromolecule #13: DNA-directed RNA polymerase subunit alpha
+Macromolecule #14: DNA-directed RNA polymerase subunit beta'
+Macromolecule #15: DNA-directed RNA polymerase subunit omega
+Macromolecule #16: 30S ribosomal protein S18
+Macromolecule #18: 30S ribosomal protein S20
+Macromolecule #19: 30S ribosomal protein S21
+Macromolecule #20: 30S ribosomal protein S2
+Macromolecule #21: 30S ribosomal protein S1
+Macromolecule #22: 30S ribosomal protein S3
+Macromolecule #23: 30S ribosomal protein S4
+Macromolecule #24: 30S ribosomal protein S5
+Macromolecule #25: 30S ribosomal protein S6
+Macromolecule #26: 30S ribosomal protein S7
+Macromolecule #27: 30S ribosomal protein S8
+Macromolecule #28: 30S ribosomal protein S9
+Macromolecule #29: 30S ribosomal protein S10
+Macromolecule #30: 30S ribosomal protein S11
+Macromolecule #31: 30S ribosomal protein S12
+Macromolecule #32: 30S ribosomal protein S14
+Macromolecule #33: 30S ribosomal protein S15
+Macromolecule #34: 30S ribosomal protein S16
+Macromolecule #35: 30S ribosomal protein S17
+Macromolecule #36: 30S ribosomal protein S19
+Macromolecule #37: 30S ribosomal protein S13
+Macromolecule #38: 50S ribosomal protein L11
+Macromolecule #39: 50S ribosomal protein L7/L12
+Macromolecule #41: 50S ribosomal protein L27
+Macromolecule #42: 50S ribosomal protein L28
+Macromolecule #44: 50S ribosomal protein L29
+Macromolecule #45: 50S ribosomal protein L30
+Macromolecule #46: 50S ribosomal protein L31
+Macromolecule #47: 50S ribosomal protein L2
+Macromolecule #48: 50S ribosomal protein L32
+Macromolecule #49: 50S ribosomal protein L3
+Macromolecule #50: 50S ribosomal protein L33
+Macromolecule #51: 50S ribosomal protein L4
+Macromolecule #52: 50S ribosomal protein L34
+Macromolecule #53: 50S ribosomal protein L5
+Macromolecule #54: 50S ribosomal protein L35
+Macromolecule #55: 50S ribosomal protein L6
+Macromolecule #56: 50S ribosomal protein L36
+Macromolecule #57: 50S ribosomal protein L9
+Macromolecule #58: 50S ribosomal protein L13
+Macromolecule #59: 50S ribosomal protein L14
+Macromolecule #60: 50S ribosomal protein L15
+Macromolecule #61: 50S ribosomal protein L16
+Macromolecule #62: 50S ribosomal protein L17
+Macromolecule #63: 50S ribosomal protein L18
+Macromolecule #64: 50S ribosomal protein L19
+Macromolecule #65: 50S ribosomal protein L20
+Macromolecule #6: NT DNA
+Macromolecule #7: T DNA
+Macromolecule #8: mRNA with 24 nt long spacer
+Macromolecule #10: E-site and P-site fMet-tRNA
+Macromolecule #17: 16S rRNA
+Macromolecule #40: 23S rRNA
+Macromolecule #43: 5S rRNA
+Macromolecule #66: MAGNESIUM ION
+Macromolecule #67: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 28.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.25 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.2 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 35602 |
| Initial angle assignment | Type: COMMON LINE |
| Final angle assignment | Type: COMMON LINE |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)