[English] 日本語
 Yorodumi
Yorodumi- EMDB-40220: 96-nm repeat unit of doublet microtubules from Chlamydomonas rein... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 96-nm repeat unit of doublet microtubules from Chlamydomonas reinhardtii flagella | |||||||||
 Map data Map data | composite map of the 96 nm repeat from C. reinhardtii | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cilia / microtubule / dynein / MOTOR PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationaxonemal central pair / axonemal doublet microtubule / radial spoke head / intraciliary transport particle B / radial spoke / axonemal dynein complex assembly / organelle / positive regulation of cilium-dependent cell motility / inner dynein arm / axonemal dynein complex ...axonemal central pair / axonemal doublet microtubule / radial spoke head / intraciliary transport particle B / radial spoke / axonemal dynein complex assembly / organelle / positive regulation of cilium-dependent cell motility / inner dynein arm / axonemal dynein complex / outer dynein arm / outer dynein arm assembly / cilium-dependent cell motility / inner dynein arm assembly / regulation of cilium beat frequency involved in ciliary motility / establishment of protein localization to organelle / 9+2 motile cilium / cilium movement involved in cell motility / intraciliary transport / axoneme assembly / dynein light chain binding / cilium movement / dynein heavy chain binding / motile cilium assembly / axonemal microtubule / cilium organization / blue light photoreceptor activity / dynein axonemal particle / negative regulation of microtubule depolymerization / nucleoside-diphosphate kinase / cell projection organization / Oxidoreductases / Set1C/COMPASS complex / UTP biosynthetic process / ciliary plasm / CTP biosynthetic process / dynein complex / myosin II complex / microtubule motor activity / microtubule associated complex / minus-end-directed microtubule motor activity / nucleoside diphosphate kinase activity / dynein light intermediate chain binding / cytoplasmic dynein complex / motile cilium / GTP biosynthetic process / microtubule-based movement / ciliary base / dynein intermediate chain binding / dynein complex binding / mitotic cytokinesis / cilium assembly / axoneme / alpha-tubulin binding / microtubule-based process / beta-tubulin binding / enzyme regulator activity / FAD binding / Hsp70 protein binding / acrosomal vesicle / GTPase activator activity / regulation of microtubule cytoskeleton organization / mitotic spindle organization / meiotic cell cycle / cell projection / peptidylprolyl isomerase / peptidyl-prolyl cis-trans isomerase activity / cell motility / ATP-dependent protein folding chaperone / Hsp90 protein binding / structural constituent of cytoskeleton / small GTPase binding / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / mitotic spindle / unfolded protein binding / protein folding / protein-folding chaperone binding / spermatogenesis / microtubule binding / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / microtubule / oxidoreductase activity / cell differentiation / cytoskeleton / calmodulin binding / cilium / ciliary basal body / hydrolase activity / cell division / GTPase activity / calcium ion binding / GTP binding / ATP hydrolysis activity / ATP binding / metal ion binding / identical protein binding / nucleus / membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Walton T / Brown A | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Axonemal structures reveal mechanoregulatory and disease mechanisms. Authors: Travis Walton / Miao Gui / Simona Velkova / Mahmoud R Fassad / Robert A Hirst / Eric Haarman / Christopher O'Callaghan / Mathieu Bottier / Thomas Burgoyne / Hannah M Mitchison / Alan Brown /      Abstract: Motile cilia and flagella beat rhythmically on the surface of cells to power the flow of fluid and to enable spermatozoa and unicellular eukaryotes to swim. In humans, defective ciliary motility can ...Motile cilia and flagella beat rhythmically on the surface of cells to power the flow of fluid and to enable spermatozoa and unicellular eukaryotes to swim. In humans, defective ciliary motility can lead to male infertility and a congenital disorder called primary ciliary dyskinesia (PCD), in which impaired clearance of mucus by the cilia causes chronic respiratory infections. Ciliary movement is generated by the axoneme, a molecular machine consisting of microtubules, ATP-powered dynein motors and regulatory complexes. The size and complexity of the axoneme has so far prevented the development of an atomic model, hindering efforts to understand how it functions. Here we capitalize on recent developments in artificial intelligence-enabled structure prediction and cryo-electron microscopy (cryo-EM) to determine the structure of the 96-nm modular repeats of axonemes from the flagella of the alga Chlamydomonas reinhardtii and human respiratory cilia. Our atomic models provide insights into the conservation and specialization of axonemes, the interconnectivity between dyneins and their regulators, and the mechanisms that maintain axonemal periodicity. Correlated conformational changes in mechanoregulatory complexes with their associated axonemal dynein motors provide a mechanism for the long-hypothesized mechanotransduction pathway to regulate ciliary motility. Structures of respiratory-cilia doublet microtubules from four individuals with PCD reveal how the loss of individual docking factors can selectively eradicate periodically repeating structures. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
-Validation report
| Summary document |  emd_40220_validation.pdf.gz emd_40220_validation.pdf.gz | 515.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40220_full_validation.pdf.gz emd_40220_full_validation.pdf.gz | 514.8 KB | Display | |
| Data in XML |  emd_40220_validation.xml.gz emd_40220_validation.xml.gz | 4.3 KB | Display | |
| Data in CIF |  emd_40220_validation.cif.gz emd_40220_validation.cif.gz | 4.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40220 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40220 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40220 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40220 | HTTPS FTP |
-Related structure data
| Related structure data |  8glvMC  8j07C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40220.map.gz / Format: CCP4 / Size: 1.3 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40220.map.gz / Format: CCP4 / Size: 1.3 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | composite map of the 96 nm repeat from C. reinhardtii | ||||||||||||||||||||||||||||||||||||
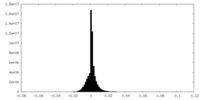
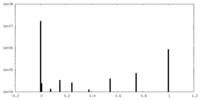
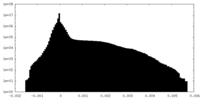
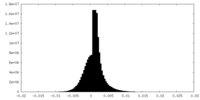
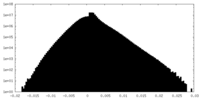
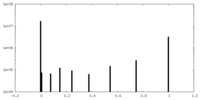
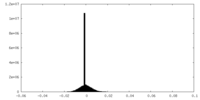
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||
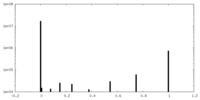
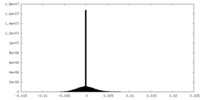
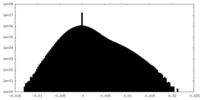
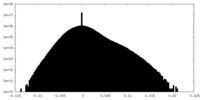
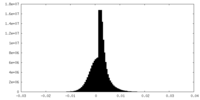
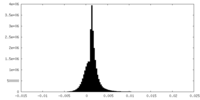
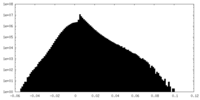
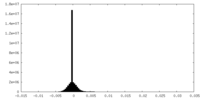
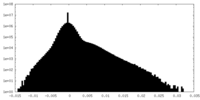
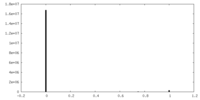
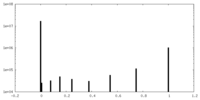
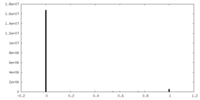
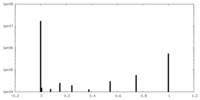
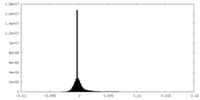
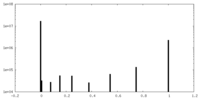
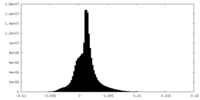
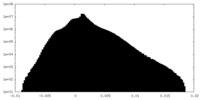
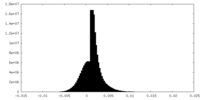
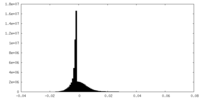
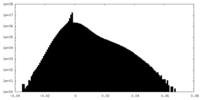
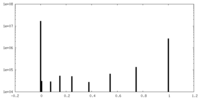
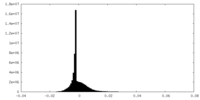
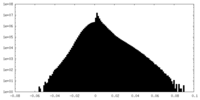
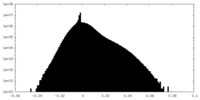
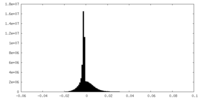
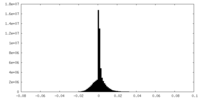
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
+Additional map: position 1 of DMT - focused refinement 1 - postprocess
+Additional map: position 1 of DMT - focused refinement 3 - postprocess
+Additional map: position 4 of DMT - focused refinement 4 - postprocess
+Additional map: position 4 of DMT - focused refinement 4 - mask
+Additional map: position 4 of DMT - focused refinement 3 - postprocess
+Additional map: position 4 of DMT - focused refinement 3 - half map 1
+Additional map: position 4 of DMT - focused refinement 3 - half map 2
+Additional map: position 4 of DMT - focused refinement 4 - half map 1
+Additional map: position 4 of DMT - focused refinement 1 - mask
+Additional map: position 4 of DMT - focused refinement 3 - mask
+Additional map: position 4 of DMT - focused refinement 1 - postprocess
+Additional map: position 4 of DMT - focused refinement 2 - postprocess
+Additional map: position 1 of DMT - focused refinement 3 - half map 2
+Additional map: position 4 of DMT - focused refinement 2 - half map 1
+Additional map: position 4 of DMT - focused refinement 2 - half map 2
+Additional map: position 4 of DMT - focused refinement 2 - mask
+Additional map: position 4 of DMT - focused refinement 6 - half map 2
+Additional map: position 4 of DMT - focused refinement 7 - half map 1
+Additional map: position 4 of DMT - focused refinement 6 - postprocess
+Additional map: position 4 of DMT - focused refinement 5 - postprocess
+Additional map: position 4 of DMT - focused refinement 5 - mask
+Additional map: position 4 of DMT - focused refinement 7 - mask
+Additional map: position 4 of DMT - focused refinement 7 - postprocess
+Additional map: position 1 of DMT - focused refinement 3 - mask
+Additional map: position 4 of DMT - focused refinement 8 - half map 1
+Additional map: position 4 of DMT - focused refinement 5 - half map 1
+Additional map: position 4 of DMT - focused refinement 8 - half map 2
+Additional map: position 4 of DMT - focused refinement 6 - half map 1
+Additional map: position 4 of DMT - focused refinement 8 - mask
+Additional map: position 4 of DMT - focused refinement 8 - postprocess
+Additional map: position 4 of DMT - focused refinement 5 - half map 2
+Additional map: position 4 of DMT - focused refinement 7 - half map 2
+Additional map: position 4 of DMT - focused refinement 6 - mask
+Additional map: NDRC closed conformation consensus - postprocess
+Additional map: position 1 of DMT - focused refinement 4 - postprocess
+Additional map: N-DRC baseplate - focused refinement 3 - postprocess
+Additional map: ODA1 core connection with IDAf - half map 2
+Additional map: ODA1 core connection with IDAf - mask
+Additional map: N-DRC linker - distal lobe - half map 2
+Additional map: NDRC closed conformation consensus - half map 1
+Additional map: N-DRC linker - central region - mask
+Additional map: N-DRC baseplate - focused refinement 1 - half map 1
+Additional map: N-DRC linker - proximal lobe - mask
+Additional map: ODA beta heavy chain motor - half map 2
+Additional map: ODA beta heavy chain motor - half map 1
+Additional map: position 1 of DMT - focused refinement 4 - half map 2
+Additional map: N-DRC baseplate - focused refinement 2 - half map 2
+Additional map: ODA2 core connection with MIA - half map 2
+Additional map: N-DRC linker - distal lobe - postprocess
+Additional map: N-DRC baseplate - focused refinement 3 - mask
+Additional map: ODA beta heavy chain motor - mask
+Additional map: ODA2 core connection with MIA - mask
+Additional map: NDRC open conformation consensus - half map 1
+Additional map: NDRC open conformation - linker - mask
+Additional map: N-DRC linker - central region - half map 1
+Additional map: N-DRC linker - proximal lobe - half map 2
+Additional map: position 1 of DMT - focused refinement 4 - mask
+Additional map: N-DRC baseplate - focused refinement 1 - half map 2
+Additional map: ODA3 core connection with N-DRC - half map 2
+Additional map: ODA3 core connection with N-DRC - mask
+Additional map: N-DRC linker - distal lobe - half map 1
+Additional map: NDRC closed conformation consensus - mask
+Additional map: N-DRC linker - proximal lobe - half map 1
+Additional map: N-DRC linker - proximal lobe - postprocess
+Additional map: N-DRC baseplate - focused refinement 2 - half map 1
+Additional map: NDRC open conformation consensus - mask
+Additional map: N-DRC baseplate - focused refinement 2 - postprocess
+Additional map: position 1 of DMT - focused refinement 3 - half map 1
+Additional map: N-DRC baseplate - focused refinement 3 - half map 1
+Additional map: NDRC closed conformation consensus - half map 2
+Additional map: N-DRC baseplate - focused refinement 2 - mask
+Additional map: N-DRC baseplate - focused refinement 1 - postprocess
+Additional map: N-DRC linker - central region - postprocess
+Additional map: N-DRC linker - distal lobe - mask
+Additional map: N-DRC baseplate - focused refinement 3 - half map 2
+Additional map: ODA3 core connection with N-DRC - postprocess
+Additional map: NDRC open conformation - linker - half map 1
+Additional map: NDRC open conformation - linker - half map 2
+Additional map: position 1 of DMT - focused refinement 5 - half map 2
+Additional map: N-DRC linker - central region - half map 2
+Additional map: N-DRC baseplate - focused refinement 1 - mask
+Additional map: ODA2 core connection with MIA - postprocess
+Additional map: ODA3 core connection with N-DRC - half map 1
+Additional map: NDRC open conformation consensus - half map 2
+Additional map: ODA beta heavy chain motor - postprocess
+Additional map: NDRC open conformation - linker - postprocess
+Additional map: ODA1 core connection with IDAf - postprocess
+Additional map: ODA2 core connection with MIA - half map 1
+Additional map: ODA1 core connection with IDAf - half map 1
+Additional map: position 1 of DMT - focused refinement 7 - half map 2
+Additional map: NDRC open conformation consensus - postprocess
+Additional map: Tetherhead complex connection with inner junction - half map 1
+Additional map: Tetherhead complex connection with inner junction - postprocess
+Additional map: IDAa base - postprocess
+Additional map: Tetherhead complex connection with IDAf motors - mask
+Additional map: IDAa base - half map 1
+Additional map: Tetherhead complex connection with IDAf motors - half map 1
+Additional map: IDAa base - half map 2
+Additional map: Tetherhead complex connection with IDAf motors - half map 2
+Additional map: Tetherhead complex connection with inner junction - half map 2
+Additional map: position 1 of DMT - focused refinement 6 - postprocess
+Additional map: IDAa base - mask
+Additional map: Tetherhead complex connection with IDAf motors - postprocess
+Additional map: Tetherhead complex connection with inner junction - mask
+Additional map: IDAf distal half - focused refinement 2 - half map 2
+Additional map: MIA / FAP57 connection - half map 1
+Additional map: IDAa motor - postprocess
+Additional map: MIA / FAP57 connection - half map 2
+Additional map: IDAc - half map 1
+Additional map: IDAa motor - half map 2
+Additional map: IDAa motor - mask
+Additional map: position 1 of DMT - focused refinement 1 - half map 1
+Additional map: position 1 of DMT - focused refinement 6 - half map 2
+Additional map: IDAf IC/LC region - half map 2
+Additional map: MIA / FAP57 connection - mask
+Additional map: IDAb - half map 2
+Additional map: MIA / FAP189 connection - mask
+Additional map: IDAc - postprocess
+Additional map: IDAf distal half - focused refinement 2 - half map 1
+Additional map: MIA / FAP57 connection - postprocess
+Additional map: IDAd motor - half map 1
+Additional map: IDAd base with RS3S - half map 1
+Additional map: IDAc - mask
+Additional map: position 1 of DMT - focused refinement 8 - postprocess
+Additional map: IDAe - half map 1
+Additional map: MIA / FAP189 connection - half map 2
+Additional map: IDAf distal half - focused refinement 1 - mask
+Additional map: IDAb - mask
+Additional map: IDAf distal half - focused refinement 1 - postprocess
+Additional map: MIA / FAP189 connection - postprocess
+Additional map: IDAf distal half - focused refinement 1 - half map 1
+Additional map: IDAe - mask
+Additional map: IDAb - postprocess
+Additional map: IDAf distal half - focused refinement 2 - postprocess
+Additional map: position 1 of DMT - focused refinement 5 - postprocess
+Additional map: MIA / FAP189 connection - half map 1
+Additional map: IDAf distal half - focused refinement 2 - mask
+Additional map: IDAd motor - mask
+Additional map: IDAd base with RS3S - postprocess
+Additional map: IDAf IC/LC region - half map 1
+Additional map: IDAe - postprocess
+Additional map: IDAd base with RS3S - half map 2
+Additional map: IDAb - half map 1
+Additional map: IDAc - half map 2
+Additional map: IDAe - half map 2
+Additional map: position 1 of DMT - focused refinement 8 - mask
+Additional map: IDAa motor - half map 1
+Additional map: IDAd base with RS3S - mask
+Additional map: IDAf IC/LC region - mask
+Additional map: IDAd motor - postprocess
+Additional map: IDAd motor - half map 2
+Additional map: IDAf IC/LC region - postprocess
+Additional map: IDAf distal half - focused refinement 1 - half map 2
+Additional map: IDAg motor - half map 1
+Additional map: IDAg base - postprocess
+Additional map: distal protrusion - half map 1
+Additional map: position 1 of DMT - focused refinement 6 - mask
+Additional map: IDAf closed conformation - core - half map 1
+Additional map: IDAf open conformation - motors - postprocess
+Additional map: IDAf open conformation - core - half map 2
+Additional map: IDAf closed conformation - motors - half map 2
+Additional map: IDAf closed conformation - motors - mask
+Additional map: IDAf open conformation - core - postprocess
+Additional map: IDAf open conformation - motors - mask
+Additional map: IDAf open conformation - core - half map 1
+Additional map: IDAf open conformation - core - mask
+Additional map: IDAg base - half map 1
+Additional map: position 1 of DMT - focused refinement 8 - half map 2
+Additional map: IDAf closed conformation - motors - half map 1
+Additional map: distal protrusion - mask
+Additional map: IDAf closed conformation - core - postprocess
+Additional map: IDAf open conformation - motors - half map 2
+Additional map: IDAg base - mask
+Additional map: IDAf open conformation - motors - half map 1
+Additional map: IDAf closed conformation - motors - postprocess
+Additional map: IDAf closed conformation - core - mask
+Additional map: IDAg base - half map 2
+Additional map: IDAg base - half map 2
+Additional map: position 1 of DMT - focused refinement 7 - mask
+Additional map: IDAf closed conformation - core - half map 2
+Additional map: distal protrusion - half map 2
+Additional map: distal protrusion - postprocess
+Additional map: IDAg base - mask
+Additional map: IDAg base - postprocess
+Additional map: position 1 of DMT - focused refinement 6 - half map 1
+Additional map: position 1 of DMT - focused refinement 7 - half map 1
+Additional map: position 1 of DMT - focused refinement 5 - half map 1
+Additional map: position 1 of DMT - focused refinement 2 - postprocess
+Additional map: position 1 of DMT - focused refinement 5 - mask
+Additional map: position 1 of DMT - focused refinement 7 - postprocess
+Additional map: position 1 of DMT - focused refinement 8 - half map 1
+Additional map: position 2 of DMT - focused refinement 1 - postprocess
+Additional map: position 2 of DMT - focused refinement 1 - half map 1
+Additional map: position 2 of DMT - focused refinement 1 - mask
+Additional map: position 2 of DMT - focused refinement 1 - half map 2
+Additional map: position 2 of DMT - focused refinement 4 - postprocess
+Additional map: position 2 of DMT - focused refinement 4 - mask
+Additional map: position 2 of DMT - focused refinement 4 - half map 2
+Additional map: position 1 of DMT - focused refinement 1 - mask
+Additional map: position 2 of DMT - focused refinement 3 - half map 2
+Additional map: position 2 of DMT - focused refinement 3 - half map 1
+Additional map: position 2 of DMT - focused refinement 3 - mask
+Additional map: position 2 of DMT - focused refinement 2 - mask
+Additional map: position 2 of DMT - focused refinement 2 - postprocess
+Additional map: position 2 of DMT - focused refinement 4 - half map 1
+Additional map: position 2 of DMT - focused refinement 2 - half map 1
+Additional map: position 2 of DMT - focused refinement 2 - half map 2
+Additional map: position 2 of DMT - focused refinement 3 - postprocess
+Additional map: position 2 of DMT - focused refinement 7 - mask
+Additional map: position 1 of DMT - focused refinement 2 - half map 2
+Additional map: position 2 of DMT - focused refinement 8 - postprocess
+Additional map: position 2 of DMT - focused refinement 8 - half map 2
+Additional map: position 2 of DMT - focused refinement 7 - half map 1
+Additional map: position 2 of DMT - focused refinement 5 - half map 1
+Additional map: position 2 of DMT - focused refinement 5 - half map 2
+Additional map: position 2 of DMT - focused refinement 8 - half map 1
+Additional map: position 2 of DMT - focused refinement 5 - mask
+Additional map: position 2 of DMT - focused refinement 6 - half map 1
+Additional map: position 2 of DMT - focused refinement 6 - half map 2
+Additional map: position 2 of DMT - focused refinement 6 - postprocess
+Additional map: position 1 of DMT - focused refinement 2 - half map 1
+Additional map: position 2 of DMT - focused refinement 7 - half map 2
+Additional map: position 2 of DMT - focused refinement 7 - postprocess
+Additional map: position 2 of DMT - focused refinement 8 - mask
+Additional map: position 2 of DMT - focused refinement 6 - mask
+Additional map: position 2 of DMT - focused refinement 5 - postprocess
+Additional map: position 3 of DMT - focused refinement 4 - mask
+Additional map: position 3 of DMT - focused refinement 3 - half map 1
+Additional map: position 3 of DMT - focused refinement 3 - postprocess
+Additional map: position 3 of DMT - focused refinement 3 - mask
+Additional map: position 3 of DMT - focused refinement 1 - postprocess
+Additional map: position 1 of DMT - focused refinement 1 - half map 2
+Additional map: position 3 of DMT - focused refinement 3 - half map 2
+Additional map: position 3 of DMT - focused refinement 2 - mask
+Additional map: position 3 of DMT - focused refinement 4 - postprocess
+Additional map: position 3 of DMT - focused refinement 4 - half map 1
+Additional map: position 3 of DMT - focused refinement 4 - half map 2
+Additional map: position 3 of DMT - focused refinement 2 - postprocess
+Additional map: position 3 of DMT - focused refinement 1 - mask
+Additional map: position 3 of DMT - focused refinement 2 - half map 1
+Additional map: position 3 of DMT - focused refinement 1 - half map 2
+Additional map: position 3 of DMT - focused refinement 1 - half map 1
+Additional map: position 1 of DMT - focused refinement 2 - mask
+Additional map: position 3 of DMT - focused refinement 2 - half map 2
+Additional map: position 3 of DMT - focused refinement 7 - mask
+Additional map: position 3 of DMT - focused refinement 7 - postprocess
+Additional map: position 3 of DMT - focused refinement 5 - mask
+Additional map: position 3 of DMT - focused refinement 6 - postprocess
+Additional map: position 3 of DMT - focused refinement 6 - mask
+Additional map: position 3 of DMT - focused refinement 8 - mask
+Additional map: position 3 of DMT - focused refinement 8 - postprocess
+Additional map: position 3 of DMT - focused refinement 5 - postprocess
+Additional map: position 3 of DMT - focused refinement 7 - half map 1
+Additional map: position 1 of DMT - focused refinement 4 - half map 1
+Additional map: position 3 of DMT - focused refinement 5 - half map 1
+Additional map: position 3 of DMT - focused refinement 8 - half map 2
+Additional map: position 3 of DMT - focused refinement 8 - half map 1
+Additional map: position 3 of DMT - focused refinement 7 - half map 1
+Additional map: position 3 of DMT - focused refinement 5 - half map 2
+Additional map: position 3 of DMT - focused refinement 6 - half map 1
+Additional map: position 3 of DMT - focused refinement 6 - half map 2
+Additional map: position 4 of DMT - focused refinement 1 - half map 1
+Additional map: position 4 of DMT - focused refinement 4 - half map 2
+Additional map: position 4 of DMT - focused refinement 1 - half map 2
- Sample components
Sample components
+Entire : 96-nm repeat unit of doublet microtubules from the flagella of Ch...
+Supramolecule #1: 96-nm repeat unit of doublet microtubules from the flagella of Ch...
+Macromolecule #1: Tubulin beta
+Macromolecule #2: Tubulin alpha
+Macromolecule #3: PACRG
+Macromolecule #4: Cilia- and flagella-associated protein 20
+Macromolecule #5: Meiosis-specific nuclear structural protein 1
+Macromolecule #6: Flagellar associated protein
+Macromolecule #7: Coiled-coil protein associated with protofilament ribbons
+Macromolecule #8: Cilia- and flagella-associated protein 45
+Macromolecule #9: FAP34
+Macromolecule #10: Flagellar associated protein
+Macromolecule #11: Protein Flattop homolog
+Macromolecule #12: FAP52
+Macromolecule #13: Nucleoside diphosphate kinase
+Macromolecule #14: Flagellar associated protein
+Macromolecule #15: Flagellar associated protein
+Macromolecule #16: Flagellar associated protein
+Macromolecule #17: FAP143
+Macromolecule #18: Flagellar associated protein
+Macromolecule #19: FAP166
+Macromolecule #20: Flagellar associated protein
+Macromolecule #21: FAP339
+Macromolecule #22: Protofilament ribbon protein of flagellar microtubules
+Macromolecule #23: Flagellar associated protein
+Macromolecule #24: Flagellar associated protein
+Macromolecule #25: Flagellar associated protein
+Macromolecule #26: Flagellar associated protein
+Macromolecule #27: FAP129
+Macromolecule #28: FAP21
+Macromolecule #29: Flagellar associated protein
+Macromolecule #30: Flagellar associated protein
+Macromolecule #31: FAP306
+Macromolecule #32: Flagellar associated protein
+Macromolecule #33: Flagellar associated protein
+Macromolecule #34: Dynein axonemal light chain 1
+Macromolecule #35: Outer dynein arm-docking complex subunit 1
+Macromolecule #36: Outer dynein arm-docking complex protein DC3
+Macromolecule #37: Dynein gamma chain, flagellar outer arm
+Macromolecule #38: IC97/Casc1 N-terminal domain-containing protein
+Macromolecule #39: Axonemal inner arm I1 intermediate chain dynein IC138
+Macromolecule #40: IC140
+Macromolecule #41: Axonemal inner arm dynein I1/f subunit
+Macromolecule #42: Dynein light chain roadblock LC7b
+Macromolecule #43: Dynein light chain roadblock LC7a
+Macromolecule #44: Dynein 8 kDa light chain, flagellar outer arm
+Macromolecule #45: Dynein light chain Tctex1
+Macromolecule #46: Inner arm dynein light chain
+Macromolecule #47: Dynein light chain 10
+Macromolecule #48: Cilia- and flagella-associated protein 73
+Macromolecule #49: Cilia- and flagella-associated protein 100
+Macromolecule #50: Dynein, 78 kDa intermediate chain, flagellar outer arm
+Macromolecule #51: WD_REPEATS_REGION domain-containing protein
+Macromolecule #52: Flagellar outer dynein arm light chain 2
+Macromolecule #53: Dynein 18 kDa light chain, flagellar outer arm
+Macromolecule #54: Dynein 11 kDa light chain, flagellar outer arm
+Macromolecule #55: Dynein light chain 9
+Macromolecule #56: Outer dynein arm protein 1
+Macromolecule #57: AAA+ ATPase domain-containing protein
+Macromolecule #58: Dynein-1-beta heavy chain, flagellar inner arm I1 complex
+Macromolecule #59: WD_REPEATS_REGION domain-containing protein
+Macromolecule #60: Inner-arm dynein f/I1 light chain
+Macromolecule #61: Heavy chain alpha
+Macromolecule #62: Flagellar outer dynein arm heavy chain beta
+Macromolecule #63: Dynein 14 kDa light chain, flagellar outer arm
+Macromolecule #64: Dynein 16 kDa light chain, flagellar outer arm
+Macromolecule #65: FAP44
+Macromolecule #66: FAP43
+Macromolecule #67: Dynein regulatory complex protein 1
+Macromolecule #68: Dynein regulatory complex subunit 2
+Macromolecule #69: Dynein regulatory complex subunit 6
+Macromolecule #70: Dynein regulatory complex subunit 4
+Macromolecule #71: Dynein regulatory complex protein 8
+Macromolecule #72: ATPase AAA-type core domain-containing protein
+Macromolecule #73: Flagellar associated protein
+Macromolecule #74: Dynein regulatory complex subunit 3
+Macromolecule #75: Dynein regulatory complex protein 9
+Macromolecule #76: Dynein regulatory complex protein 10
+Macromolecule #77: DRC5
+Macromolecule #78: DUF4201 domain-containing protein
+Macromolecule #79: Calmodulin
+Macromolecule #80: Caltractin
+Macromolecule #81: AAA+ ATPase domain-containing protein
+Macromolecule #82: DUF4201 domain-containing protein
+Macromolecule #83: Cilia- and flagella-associated protein 299
+Macromolecule #84: Cilia- and flagella-associated protein 53
+Macromolecule #85: FAP210
+Macromolecule #86: Actin
+Macromolecule #87: Flagellar radial spoke protein 5
+Macromolecule #88: Flagellar radial spoke protein 1
+Macromolecule #89: Flagellar radial spoke protein 2
+Macromolecule #90: Radial spoke protein 3
+Macromolecule #91: Radial spoke protein 4
+Macromolecule #92: Flagellar radial spoke protein 6
+Macromolecule #93: Radial spoke protein 7
+Macromolecule #94: Radial spoke protein 9
+Macromolecule #95: Radial spoke protein 10
+Macromolecule #96: Radial spoke protein 11
+Macromolecule #97: Peptidyl-prolyl cis-trans isomerase
+Macromolecule #98: Radial spoke protein 14
+Macromolecule #99: Radial spoke protein 16
+Macromolecule #100: UNKNOWN
+Macromolecule #101: Nucleoside diphosphate kinase 6
+Macromolecule #102: Cytochrome b5 heme-binding domain-containing protein
+Macromolecule #103: FAP385
+Macromolecule #104: FAP207
+Macromolecule #105: FAP253
+Macromolecule #106: 28 kDa inner dynein arm light chain, axonemal
+Macromolecule #107: AAA+ ATPase domain-containing protein
+Macromolecule #108: UNKNOWN
+Macromolecule #109: Radial spoke protein 15
+Macromolecule #110: Radial spike protein 8
+Macromolecule #111: DHC_N2 domain-containing protein
+Macromolecule #112: Flagellar-associated protein 59
+Macromolecule #113: Flagellar-associated protein 172
+Macromolecule #114: Flagellar associated protein
+Macromolecule #115: Cilia- and flagella-associated protein 58
+Macromolecule #116: FAP251
+Macromolecule #117: Cilia- and flagella-associated protein 61
+Macromolecule #118: DHC2
+Macromolecule #119: Subunit of axonemal inner dynein
+Macromolecule #120: Subunit of axonemal inner dynein
+Macromolecule #121: CFAP91 domain-containing protein
+Macromolecule #122: AAA+ ATPase domain-containing protein
+Macromolecule #123: AAA+ ATPase domain-containing protein
+Macromolecule #124: Cilia- and flagella-associated protein 77
+Macromolecule #125: Flagellar associated protein
+Macromolecule #126: Flagellar associated protein
+Macromolecule #127: FAP1
+Macromolecule #128: GUANOSINE-5'-TRIPHOSPHATE
+Macromolecule #129: GUANOSINE-5'-DIPHOSPHATE
+Macromolecule #130: MAGNESIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 20 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV Details: 2.5 microliters of splayed axoneme sample at 16-26 mg/mL were applied to glow-discharged C-Flat 1.2/1.3-4Cu grids at 100% humidity. The samples were then incubated 10 s, blotted for 10 s, ...Details: 2.5 microliters of splayed axoneme sample at 16-26 mg/mL were applied to glow-discharged C-Flat 1.2/1.3-4Cu grids at 100% humidity. The samples were then incubated 10 s, blotted for 10 s, and plunged into liquid ethane. | |||||||||||||||
| Details | Axonemes were splayed using 10 mM ATP and 0.75 mM CaCl2 for one hour at room temperature before centrifugation and resuspension. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 36918 / Average exposure time: 3.7 sec. / Average electron dose: 61.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 64000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)