[English] 日本語
 Yorodumi
Yorodumi- EMDB-39109: SARS-CoV-2 DMV nsp3-4 pore complex (consensus-pore, C6 symmetry) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 DMV nsp3-4 pore complex (consensus-pore, C6 symmetry) | |||||||||
 Map data Map data | local resolution map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Double membrane vesicle / pore complex / nsp3 / nsp4 / RNA transport / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein guanylyltransferase activity / RNA endonuclease activity producing 3'-phosphomonoesters, hydrolytic mechanism / mRNA guanylyltransferase activity / 5'-3' RNA helicase activity / Lyases; Phosphorus-oxygen lyases / Assembly of the SARS-CoV-2 Replication-Transcription Complex (RTC) / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / Maturation of replicase proteins / TRAF3-dependent IRF activation pathway / ISG15-specific peptidase activity ...protein guanylyltransferase activity / RNA endonuclease activity producing 3'-phosphomonoesters, hydrolytic mechanism / mRNA guanylyltransferase activity / 5'-3' RNA helicase activity / Lyases; Phosphorus-oxygen lyases / Assembly of the SARS-CoV-2 Replication-Transcription Complex (RTC) / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / Maturation of replicase proteins / TRAF3-dependent IRF activation pathway / ISG15-specific peptidase activity / Transcription of SARS-CoV-2 sgRNAs / snRNP Assembly / Translation of Replicase and Assembly of the Replication Transcription Complex / Replication of the SARS-CoV-2 genome / Hydrolases; Acting on ester bonds; Exoribonucleases producing 5'-phosphomonoesters / double membrane vesicle viral factory outer membrane / SARS coronavirus main proteinase / host cell endoplasmic reticulum-Golgi intermediate compartment / 5'-3' DNA helicase activity / 3'-5'-RNA exonuclease activity / symbiont-mediated degradation of host mRNA / host cell endosome / mRNA guanylyltransferase / symbiont-mediated suppression of host ISG15-protein conjugation / symbiont-mediated suppression of host toll-like receptor signaling pathway / G-quadruplex RNA binding / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IRF3 activity / omega peptidase activity / mRNA (guanine-N7)-methyltransferase / methyltransferase cap1 / SARS-CoV-2 modulates host translation machinery / host cell Golgi apparatus / symbiont-mediated suppression of host NF-kappaB cascade / DNA helicase / symbiont-mediated perturbation of host ubiquitin-like protein modification / methyltransferase cap1 activity / ubiquitinyl hydrolase 1 / cysteine-type deubiquitinase activity / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases / single-stranded RNA binding / regulation of autophagy / viral protein processing / lyase activity / host cell perinuclear region of cytoplasm / host cell endoplasmic reticulum membrane / RNA helicase / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / copper ion binding / symbiont-mediated suppression of host gene expression / symbiont-mediated activation of host autophagy / viral translational frameshifting / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / lipid binding / host cell nucleus / SARS-CoV-2 activates/modulates innate and adaptive immune responses / ATP hydrolysis activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
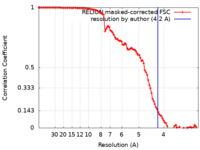
| Method | subtomogram averaging / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Huang YX / Zhong LJ / Zhang WX / Ni T | |||||||||
| Funding support |  Hong Kong, 1 items Hong Kong, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Molecular architecture of coronavirus double-membrane vesicle pore complex. Authors: Yixin Huang / Tongyun Wang / Lijie Zhong / Wenxin Zhang / Yu Zhang / Xiulian Yu / Shuofeng Yuan / Tao Ni /  Abstract: Coronaviruses remodel the intracellular host membranes during replication, forming double-membrane vesicles (DMVs) to accommodate viral RNA synthesis and modifications. SARS-CoV-2 non-structural ...Coronaviruses remodel the intracellular host membranes during replication, forming double-membrane vesicles (DMVs) to accommodate viral RNA synthesis and modifications. SARS-CoV-2 non-structural protein 3 (nsp3) and nsp4 are the minimal viral components required to induce DMV formation and to form a double-membrane-spanning pore, essential for the transport of newly synthesized viral RNAs. The mechanism of DMV pore complex formation remains unknown. Here we describe the molecular architecture of the SARS-CoV-2 nsp3-nsp4 pore complex, as resolved by cryogenic electron tomography and subtomogram averaging in isolated DMVs. The structures uncover an unexpected stoichiometry and topology of the nsp3-nsp4 pore complex comprising 12 copies each of nsp3 and nsp4, organized in 4 concentric stacking hexamer rings, mimicking a miniature nuclear pore complex. The transmembrane domains are interdigitated to create a high local curvature at the double-membrane junction, coupling double-membrane reorganization with pore formation. The ectodomains form extensive contacts in a pseudo-12-fold symmetry, belting the pore complex from the intermembrane space. A central positively charged ring of arginine residues coordinates the putative RNA translocation, essential for virus replication. Our work establishes a framework for understanding DMV pore formation and RNA translocation, providing a structural basis for the development of new antiviral strategies to combat coronavirus infection. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39109.map.gz emd_39109.map.gz | 51.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39109-v30.xml emd-39109-v30.xml emd-39109.xml emd-39109.xml | 21.2 KB 21.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_39109_fsc.xml emd_39109_fsc.xml | 10.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_39109.png emd_39109.png | 117.9 KB | ||
| Masks |  emd_39109_msk_1.map emd_39109_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-39109.cif.gz emd-39109.cif.gz | 7.8 KB | ||
| Others |  emd_39109_half_map_1.map.gz emd_39109_half_map_1.map.gz emd_39109_half_map_2.map.gz emd_39109_half_map_2.map.gz | 43.4 MB 43.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39109 http://ftp.pdbj.org/pub/emdb/structures/EMD-39109 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39109 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39109 | HTTPS FTP |
-Related structure data
| Related structure data |  8yb5MC  8yaxC  8yb7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_39109.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39109.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local resolution map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.571 Å | ||||||||||||||||||||||||||||||||||||
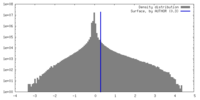
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_39109_msk_1.map emd_39109_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_39109_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_39109_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 DMV nsp3-4 pore complex
| Entire | Name: SARS-CoV-2 DMV nsp3-4 pore complex |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 DMV nsp3-4 pore complex
| Supramolecule | Name: SARS-CoV-2 DMV nsp3-4 pore complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Papain-like protease nsp3
| Macromolecule | Name: Papain-like protease nsp3 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: ubiquitinyl hydrolase 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 217.471188 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: APTKVTFGDD TVIEVQGYKS VNITFELDER IDKVLNEKCS AYTVELGTEV NEFACVVADA VIKTLQPVSE LLTPLGIDLD EWSMATYYL FDESGEFKLA SHMYCSFYPP DEDEEEGDCE EEEFEPSTQY EYGTEDDYQG KPLEFGATSA ALQPEEEQEE D WLDDDSQQ ...String: APTKVTFGDD TVIEVQGYKS VNITFELDER IDKVLNEKCS AYTVELGTEV NEFACVVADA VIKTLQPVSE LLTPLGIDLD EWSMATYYL FDESGEFKLA SHMYCSFYPP DEDEEEGDCE EEEFEPSTQY EYGTEDDYQG KPLEFGATSA ALQPEEEQEE D WLDDDSQQ TVGQQDGSED NQTTTIQTIV EVQPQLEMEL TPVVQTIEVN SFSGYLKLTD NVYIKNADIV EEAKKVKPTV VV NAANVYL KHGGGVAGAL NKATNNAMQV ESDDYIATNG PLKVGGSCVL SGHNLAKHCL HVVGPNVNKG EDIQLLKSAY ENF NQHEVL LAPLLSAGIF GADPIHSLRV CVDTVRTNVY LAVFDKNLYD KLVSSFLEMK SEKQVEQKIA EIPKEEVKPF ITES KPSVE QRKQDDKKIK ACVEEVTTTL EETKFLTENL LLYIDINGNL HPDSATLVSD IDITFLKKDA PYIVGDVVQE GVLTA VVIP TKKAGGTTEM LAKALRKVPT DNYITTYPGQ GLNGYTVEEA KTVLKKCKSA FYILPSIISN EKQEILGTVS WNLREM LAH AEETRKLMPV CVETKAIVST IQRKYKGIKI QEGVVDYGAR FYFYTSKTTV ASLINTLNDL NETLVTMPLG YVTHGLN LE EAARYMRSLK VPATVSVSSP DAVTAYNGYL TSSSKTPEEH FIETISLAGS YKDWSYSGQS TQLGIEFLKR GDKSVYYT S NPTTFHLDGE VITFDNLKTL LSLREVRTIK VFTTVDNINL HTQVVDMSMT YGQQFGPTYL DGADVTKIKP HNSHEGKTF YVLPNDDTLR VEAFEYYHTT DPSFLGRYMS ALNHTKKWKY PQVNGLTSIK WADNNCYLAT ALLTLQQIEL KFNPPALQDA YYRARAGEA ANFCALILAY CNKTVGELGD VRETMSYLFQ HANLDSCKRV LNVVCKTCGQ QQTTLKGVEA VMYMGTLSYE Q FKKGVQIP CTCGKQATKY LVQQESPFVM MSAPPAQYEL KHGTFTCASE YTGNYQCGHY KHITSKETLY CIDGALLTKS SE YKGPITD VFYKENSYTT TIKPVTYKLD GVVCTEIDPK LDNYYKKDNS YFTEQPIDLV PNQPYPNASF DNFKFVCDNI KFA DDLNQL TGYKKPASRE LKVTFFPDLN GDVVAIDYKH YTPSFKKGAK LLHKPIVWHV NNATNKATYK PNTWCIRCLW STKP VETSN SFDVLKSEDA QGMDNLACED LKPVSEEVVE NPTIQKDVLE CNVKTTEVVG DIILKPANNS LKITEEVGHT DLMAA YVDN SSLTIKKPNE LSRVLGLKTL ATHGLAAVNS VPWDTIANYA KPFLNKVVST TTNIVTRCLN RVCTNYMPYF FTLLLQ LCT FTRSTNSRIK ASMPTTIAKN TVKSVGKFCL EASFNYLKSP NFSKLINIII WFLLLSVCLG SLIYSTAALG VLMSNLG MP SYCTGYREGY LNSTNVTIAT YCTGSIPCSV CLSGLDSLDT YPSLETIQIT ISSFKWDLTA FGLVAEWFLA YILFTRFF Y VLGLAAIMQL FFSYFAVHFI SNSWLMWLII NLVQMAPISA MVRMYIFFAS FYYVWKSYVH VVDGCNSSTC MMCYKRNRA TRVECTTIVN GVRRSFYVYA NGGKGFCKLH NWNCVNCDTF CAGSTFISDE VARDLSLQFK RPINPTDQSS YIVDSVTVKN GSIHLYFDK AGQKTYERHS LSHFVNLDNL RANNTKGSLP INVIVFDGKS KCEESSAKSA SVYYSQLMCQ PILLLDQALV S DVGDSAEV AVKMFDAYVN TFSSTFNVPM EKLKTLVATA EAELAKNVSL DNVLSTFISA ARQGFVDSDV ETKDVVECLK LS HQSDIEV TGDSCNNYML TYNKVENMTP RDLGACIDCS ARHINAQVAK SHNIALIWNV KDFMSLSEQL RKQIRSAAKK NNL PFKLTC ATTRQVVNVV TTKIALKGG UniProtKB: Replicase polyprotein 1ab |
-Macromolecule #2: Non-structural protein 4
| Macromolecule | Name: Non-structural protein 4 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 56.229582 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: KIVNNWLKQL IKVTLVFLFV AAIFYLITPV HVMSKHTDFS SEIIGYKAID GGVTRDIAST DTCFANKHAD FDTWFSQRGG SYTNDKACP LIAAVITREV GFVVPGLPGT ILRTTNGDFL HFLPRVFSAV GNICYTPSKL IEYTDFATSA CVLAAECTIF K DASGKPVP ...String: KIVNNWLKQL IKVTLVFLFV AAIFYLITPV HVMSKHTDFS SEIIGYKAID GGVTRDIAST DTCFANKHAD FDTWFSQRGG SYTNDKACP LIAAVITREV GFVVPGLPGT ILRTTNGDFL HFLPRVFSAV GNICYTPSKL IEYTDFATSA CVLAAECTIF K DASGKPVP YCYDTNVLEG SVAYESLRPD TRYVLMDGSI IQFPNTYLEG SVRVVTTFDS EYCRHGTCER SEAGVCVSTS GR WVLNNDY YRSLPGVFCG VDAVNLLTNM FTPLIQPIGA LDISASIVAG GIVAIVVTCL AYYFMRFRRA FGEYSHVVAF NTL LFLMSF TVLCLTPVYS FLPGVYSVIY LYLTFYLTND VSFLAHIQWM VMFTPLVPFW ITIAYIICIS TKHFYWFFSN YLKR RVVFN GVSFSTFEEA ALCTFLLNKE MYLKLRSDVL LPLTQYNRYL ALYNKYKYFS GAMDTTSYRE AACCHLAKAL NDFSN SGSD VLYQPPQTSI TSAVLQ UniProtKB: Replicase polyprotein 1ab |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
Details: 150mM NaCl, 10mM Tris-HCl, 1mM EDTA | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: EMS Lacey Carbon / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Software | Name: SerialEM |
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Average electron dose: 3.0 e/Å2 Details: The tilt-series were acquired using Thermofisher Krios equipped with a Falcon 4i camera and Selectris energy filter. A dose-symmetric scheme (group of 2) was used, with a tilt range of -51 ...Details: The tilt-series were acquired using Thermofisher Krios equipped with a Falcon 4i camera and Selectris energy filter. A dose-symmetric scheme (group of 2) was used, with a tilt range of -51 degree to 51 degree (or -60 to 60) at 3 degree increments and an exposure dose of 3 e-/A2 per image. The total dose was 105 or 123 e-/A2. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 6.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Software | Name:  UCSF Chimera (ver. 1.16) UCSF Chimera (ver. 1.16) |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-8yb5: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)