[English] 日本語
 Yorodumi
Yorodumi- EMDB-38466: Cryo-EM structure of the RhoG/DOCK5/ELMO1/Rac1 complex: RhoG/DOCK... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the RhoG/DOCK5/ELMO1/Rac1 complex: RhoG/DOCK5/ELMO1 focused map | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | ELMO / DOCK / GEF / GTPASE / RHO / RAC / SIGNALING PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of ruffle assembly / negative regulation of vascular associated smooth muscle contraction / podosome assembly / cortical cytoskeleton organization / activation of GTPase activity / guanyl-nucleotide exchange factor complex / bone remodeling / myoblast fusion / positive regulation of vascular associated smooth muscle cell migration / Nef and signal transduction ...regulation of ruffle assembly / negative regulation of vascular associated smooth muscle contraction / podosome assembly / cortical cytoskeleton organization / activation of GTPase activity / guanyl-nucleotide exchange factor complex / bone remodeling / myoblast fusion / positive regulation of vascular associated smooth muscle cell migration / Nef and signal transduction / RHO GTPases activate KTN1 / podosome / anchoring junction / phagocytosis, engulfment / establishment or maintenance of cell polarity / Rac protein signal transduction / positive regulation of epithelial cell migration / regulation of postsynapse assembly / RHOG GTPase cycle / PTK6 Regulates RHO GTPases, RAS GTPase and MAP kinases / Rho protein signal transduction / GPVI-mediated activation cascade / RAC1 GTPase cycle / positive regulation of substrate adhesion-dependent cell spreading / GTPase activator activity / guanyl-nucleotide exchange factor activity / actin filament organization / secretory granule membrane / cell chemotaxis / regulation of actin cytoskeleton organization / positive regulation of protein localization to plasma membrane / cell projection / FCGR3A-mediated phagocytosis / cell motility / Regulation of actin dynamics for phagocytic cup formation / SH3 domain binding / VEGFA-VEGFR2 Pathway / small GTPase binding / Constitutive Signaling by Aberrant PI3K in Cancer / cell migration / PIP3 activates AKT signaling / regulation of cell shape / Factors involved in megakaryocyte development and platelet production / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / actin cytoskeleton organization / cytoplasmic vesicle / cytoskeleton / postsynapse / focal adhesion / GTPase activity / positive regulation of cell population proliferation / apoptotic process / Neutrophil degranulation / protein kinase binding / endoplasmic reticulum membrane / positive regulation of DNA-templated transcription / GTP binding / glutamatergic synapse / extracellular exosome / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
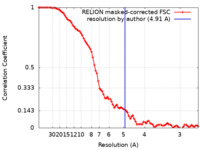
| Method | single particle reconstruction / cryo EM / Resolution: 4.91 Å | ||||||||||||
 Authors Authors | Kukimoto-Niino M / Katsura K / Ishizuka-Katsura Y / Mishima-Tsumagari C / Yonemochi M / Inoue M / Nakagawa R / Kaushik R / Zhang KYJ / Shirouzu M | ||||||||||||
| Funding support |  Japan, 3 items Japan, 3 items
| ||||||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2024 Journal: J Biol Chem / Year: 2024Title: RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state. Authors: Mutsuko Kukimoto-Niino / Kazushige Katsura / Yoshiko Ishizuka-Katsura / Chiemi Mishima-Tsumagari / Mayumi Yonemochi / Mio Inoue / Reiko Nakagawa / Rahul Kaushik / Kam Y J Zhang / Mikako Shirouzu /  Abstract: The dedicator of cytokinesis (DOCK)/engulfment and cell motility (ELMO) complex serves as a guanine nucleotide exchange factor (GEF) for the GTPase Rac. RhoG, another GTPase, activates the ELMO-DOCK- ...The dedicator of cytokinesis (DOCK)/engulfment and cell motility (ELMO) complex serves as a guanine nucleotide exchange factor (GEF) for the GTPase Rac. RhoG, another GTPase, activates the ELMO-DOCK-Rac pathway during engulfment and migration. Recent cryo-EM structures of the DOCK2/ELMO1 and DOCK2/ELMO1/Rac1 complexes have identified closed and open conformations that are key to understanding the autoinhibition mechanism. Nevertheless, the structural details of RhoG-mediated activation of the DOCK/ELMO complex remain elusive. Herein, we present cryo-EM structures of DOCK5/ELMO1 alone and in complex with RhoG and Rac1. The DOCK5/ELMO1 structure exhibits a closed conformation similar to that of DOCK2/ELMO1, suggesting a shared regulatory mechanism of the autoinhibitory state across DOCK-A/B subfamilies (DOCK1-5). Conversely, the RhoG/DOCK5/ELMO1/Rac1 complex adopts an open conformation that differs from that of the DOCK2/ELMO1/Rac1 complex, with RhoG binding to both ELMO1 and DOCK5. The alignment of the DOCK5 phosphatidylinositol (3,4,5)-trisphosphate binding site with the RhoG C-terminal lipidation site suggests simultaneous binding of RhoG and DOCK5/ELMO1 to the plasma membrane. Structural comparison of the apo and RhoG-bound states revealed that RhoG facilitates a closed-to-open state conformational change of DOCK5/ELMO1. Biochemical and surface plasmon resonance (SPR) assays confirm that RhoG enhances the Rac GEF activity of DOCK5/ELMO1 and increases its binding affinity for Rac1. Further analysis of structural variability underscored the conformational flexibility of the DOCK5/ELMO1/Rac1 complex core, potentially facilitating the proximity of the DOCK5 GEF domain to the plasma membrane. These findings elucidate the structural mechanism underlying the RhoG-induced allosteric activation and membrane binding of the DOCK/ELMO complex. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38466.map.gz emd_38466.map.gz | 49.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38466-v30.xml emd-38466-v30.xml emd-38466.xml emd-38466.xml | 25.7 KB 25.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_38466_fsc.xml emd_38466_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_38466.png emd_38466.png | 107.1 KB | ||
| Filedesc metadata |  emd-38466.cif.gz emd-38466.cif.gz | 8.4 KB | ||
| Others |  emd_38466_half_map_1.map.gz emd_38466_half_map_1.map.gz emd_38466_half_map_2.map.gz emd_38466_half_map_2.map.gz | 40.9 MB 40.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38466 http://ftp.pdbj.org/pub/emdb/structures/EMD-38466 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38466 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38466 | HTTPS FTP |
-Validation report
| Summary document |  emd_38466_validation.pdf.gz emd_38466_validation.pdf.gz | 934.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_38466_full_validation.pdf.gz emd_38466_full_validation.pdf.gz | 934 KB | Display | |
| Data in XML |  emd_38466_validation.xml.gz emd_38466_validation.xml.gz | 14.1 KB | Display | |
| Data in CIF |  emd_38466_validation.cif.gz emd_38466_validation.cif.gz | 20.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38466 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38466 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38466 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38466 | HTTPS FTP |
-Related structure data
| Related structure data |  8xm7MC  8jhkC  8zj2C  8zjiC  8zjjC  8zjkC  8zjlC  8zjmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38466.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38466.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.33 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_38466_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
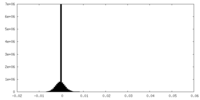
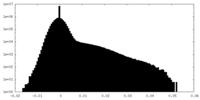
| Density Histograms |
-Half map: #2
| File | emd_38466_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
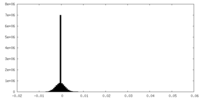
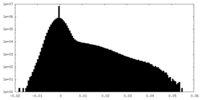
| Density Histograms |
- Sample components
Sample components
-Entire : RhoG/DOCK5/ELMO1 complex
| Entire | Name: RhoG/DOCK5/ELMO1 complex |
|---|---|
| Components |
|
-Supramolecule #1: RhoG/DOCK5/ELMO1 complex
| Supramolecule | Name: RhoG/DOCK5/ELMO1 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Engulfment and cell motility protein 1
| Macromolecule | Name: Engulfment and cell motility protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 84.337719 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GGSGGSMPPP ADIVKVAIEW PGAYPKLMEI DQKKPLSAII KEVCDGWSLA NHEYFALQHA DSSNFYITEK NRNEIKNGTI LRLTTSPAQ NAQQLHERIQ SSSMDAKLEA LKDLASLSRD VTFAQEFINL DGISLLTQMV ESGTERYQKL QKIMKPCFGD M LSFTLTAF ...String: GGSGGSMPPP ADIVKVAIEW PGAYPKLMEI DQKKPLSAII KEVCDGWSLA NHEYFALQHA DSSNFYITEK NRNEIKNGTI LRLTTSPAQ NAQQLHERIQ SSSMDAKLEA LKDLASLSRD VTFAQEFINL DGISLLTQMV ESGTERYQKL QKIMKPCFGD M LSFTLTAF VELMDHGIVS WDTFSVAFIK KIASFVNKSA IDISILQRSL AILESMVLNS HDLYQKVAQE ITIGQLIPHL QG SDQEIQT YTIAVINALF LKAPDERRQE MANILAQKQL RSIILTHVIR AQRAINNEMA HQLYVLQVLT FNLLEDRMMT KMD PQDQAQ RDIIFELRRI AFDAESEPNN SSGSMEKRKS MYTRDYKKLG FINHVNPAMD FTQTPPGMLA LDNMLYFAKH HQDA YIRIV LENSSREDKH ECPFGRSSIE LTKMLCEILK VGELPSETCN DFHPMFFTHD RSFEEFFCIC IQLLNKTWKE MRATS EDFN KVMQVVKEQV MRALTTKPSS LDQFKSKLQN LSYTEILKIR QSERMNQEDF QSRPILELKE KIQPEILELI KQQRLN RLV EGTCFRKLNA RRRQDKFWYC RLSPNHKVLH YGDLEESPQG EVPHDSLQDK LPVADIKAVV TGKDCPHMKE KGALKQN KE VLELAFSILY DSNCQLNFIA PDKHEYCIWT DGLNALLGKD MMSDLTRNDL DTLLSMEIKL RLLDLENIQI PDAPPPIP K EPSNYDFVYD CN UniProtKB: Engulfment and cell motility protein 1 |
-Macromolecule #2: Dedicator of cytokinesis protein 5
| Macromolecule | Name: Dedicator of cytokinesis protein 5 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 191.492125 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GGSGGSMARW IPTKRQKYGV AIYNYNASQD VELSLQIGDT VHILEMYEGW YRGYTLQNKS KKGIFPETYI HLKEATVEDL GQHETVIPG ELPLVQELTS TLREWAVIWR KLYVNNKLTL FRQLQQMTYS LIEWRSQILS GTLPKDELAE LKKKVTAKID H GNRMLGLD ...String: GGSGGSMARW IPTKRQKYGV AIYNYNASQD VELSLQIGDT VHILEMYEGW YRGYTLQNKS KKGIFPETYI HLKEATVEDL GQHETVIPG ELPLVQELTS TLREWAVIWR KLYVNNKLTL FRQLQQMTYS LIEWRSQILS GTLPKDELAE LKKKVTAKID H GNRMLGLD LVVRDDNGNI LDPDETSTIA LFKAHEVASK RIEEKIQEEK SILQNLDLRG QSIFSTIHTY GLYVNFKNFV CN IGEDAEL FMALYDPDQS TFISENYLIR WGSNGMPKEI EKLNNLQAVF TDLSSMDLIR PRVSLVCQIV RVGHMELKEG KKH TCGLRR PFGVAVMDIT DIIHGKVDDE EKQHFIPFQQ IAMETYIRQR QLIMSPLITS HVIGENEPLT SVLNKVIAAK EVNH KGQGL WVSLKLLPGD LTQVQKNFSH LVDRSTAIAR KMGFPEIILP GDVRNDIYVT LIHGEFDKGK KKTPKNVEVT MSVHD EEGK LLEKAIHPGA GYEGISEYKS VVYYQVKQPC WYETVKVSIA IEEVTRCHIR FTFRHRSSQE TRDKSERAFG VAFVKL MNP DGTTLQDGRH DLVVYKGDNK KMEDAKFYLT LPGTKMEMEE KELQASKNLV TFTPSKDSTK DSFQIATLIC STKLTQN VD LLGLLNWRSN SQNIKHNLKK LMEVDGGEIV KFLQDTLDAL FNIMMEMSDS ETYDFLVFDA LVFIISLIGD IKFQHFNP V LETYIYKHFS ATLAYVKLSK VLNFYVANAD DSSKTELLFA ALKALKYLFR FIIQSRVLYL RFYGQSKDGD EFNNSIRQL FLAFNMLMDR PLEEAVKIKG AALKYLPSII NDVKLVFDPV ELSVLFCKFI QSIPDNQLVR QKLNCMTKIV ESTLFRQSEC REVLLPLLT DQLSGQLDDN SNKPDHEASS QLLSNILEVL DRKDVGATAV HIQLIMERLL RRINRTVIGM NRQSPHIGSF V ACMIALLQ QMDDSHYSHY ISTFKTRQDI IDFLMETFIM FKDLIGKNVY AKDWMVMNMT QNRVFLRAIN QFAEVLTRFF MD QASFELQ LWNNYFHLAV AFLTHESLQL ETFSQAKRNK IVKKYGDMRK EIGFRIRDMW YNLGPHKIKF IPSMVGPILE VTL TPEVEL RKATIPIFFD MMQCEFNFSG NGNFHMFENE LITKLDQEVE GGRGDEQYKV LLEKLLLEHC RKHKYLSSSG EVFA LLVSS LLENLLDYRT IIMQDESKEN RMSCTVNVLN FYKEKKREDI YIRYLYKLRD LHRDCENYTE AAYTLLLHAE LLQWS DKPC VPHLLQRDSY YVYTQQELKE KLYQEIISYF DKGKMWEKAI KLSKELAETY ESKVFDYEGL GNLLKKRASF YENIIK AMR PQPEYFAVGY YGQGFPSFLR NKIFIYRGKE YERREDFSLR LLTQFPNAEK MTSTTPPGED IKSSPKQYMQ CFTVKPV MS LPPSYKDKPV PEQILNYYRA NEVQQFRYSR PFRKGEKDPD NEFATMWIER TTYTTAYTFP GILKWFEVKQ ISTEEISP L ENAIETMELT NERISNCVQQ HAWDRSLSVH PLSMLLSGIV DPAVMGGFSN YEKAFFTEKY LQEHPEDQEK VELLKRLIA LQMPLLTEGI RIHGEKLTEQ LKPLHERLSS CFRELKEKVE KHYGVITL UniProtKB: Dedicator of cytokinesis protein 5 |
-Macromolecule #3: Rho-related GTP-binding protein RhoG
| Macromolecule | Name: Rho-related GTP-binding protein RhoG / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.362359 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSSGSSGMQS IKCVVVGDGA VGKTCLLICY TTNAFPKEYI PTVFDNYSAQ SAVDGRTVNL NLWDTAGLEE YDRLRTLSYP QTNVFVICF SIASPPSYEN VRHKWHPEVC HHCPDVPILL VGTKKDLRAQ PDTLRRLKEQ GQAPITPQQG QALAKQIHAV R YLECSALQ ...String: GSSGSSGMQS IKCVVVGDGA VGKTCLLICY TTNAFPKEYI PTVFDNYSAQ SAVDGRTVNL NLWDTAGLEE YDRLRTLSYP QTNVFVICF SIASPPSYEN VRHKWHPEVC HHCPDVPILL VGTKKDLRAQ PDTLRRLKEQ GQAPITPQQG QALAKQIHAV R YLECSALQ QDGVKEVFAE AVRAVLNPTP IKRSGPSSGE NLYFQ UniProtKB: Rho-related GTP-binding protein RhoG |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 1 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 11976 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 64000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Output model |  PDB-8xm7: |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)