[English] 日本語
 Yorodumi
Yorodumi- EMDB-36996: Cryo EM structure of the products-bound PGAP1(Bst1)-H443N from Ch... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo EM structure of the products-bound PGAP1(Bst1)-H443N from Chaetomium thermophilum | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Bst1 / Glycosylphosphatidylinositol / GPI anchoring / GPI-AP / GPI-AP remodelase / Integral membrane enzyme / Lipase / Lipid remodeling / Membrane enzyme / Membrane protein / Nanodisc / PGAP1 / TGP / Thermostable green fluorescence protein / Transmembrane enzyme / Triad enzyme. | |||||||||
| Function / homology |  Function and homology information Function and homology informationGPI anchor metabolic process / phosphatidylinositol deacylase activity / regulation of lipopolysaccharide-mediated signaling pathway / negative regulation of complement activation / regulation of complement-dependent cytotoxicity / regulation of complement activation / respiratory burst / positive regulation of CD4-positive, alpha-beta T cell activation / positive regulation of CD4-positive, alpha-beta T cell proliferation / Class B/2 (Secretin family receptors) ...GPI anchor metabolic process / phosphatidylinositol deacylase activity / regulation of lipopolysaccharide-mediated signaling pathway / negative regulation of complement activation / regulation of complement-dependent cytotoxicity / regulation of complement activation / respiratory burst / positive regulation of CD4-positive, alpha-beta T cell activation / positive regulation of CD4-positive, alpha-beta T cell proliferation / Class B/2 (Secretin family receptors) / ficolin-1-rich granule membrane / complement activation, classical pathway / endoplasmic reticulum to Golgi vesicle-mediated transport / transport vesicle / side of membrane / COPI-mediated anterograde transport / endoplasmic reticulum-Golgi intermediate compartment membrane / secretory granule membrane / Regulation of Complement cascade / bioluminescence / generation of precursor metabolites and energy / positive regulation of T cell cytokine production / protein transport / positive regulation of cytosolic calcium ion concentration / virus receptor activity / Hydrolases; Acting on ester bonds / membrane raft / Golgi membrane / innate immune response / Neutrophil degranulation / lipid binding / endoplasmic reticulum membrane / cell surface / extracellular exosome / extracellular region / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) / Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) /  Psychromonas sp. B3M02 (bacteria) / Psychromonas sp. B3M02 (bacteria) /  Homo sapiens (human) Homo sapiens (human) | |||||||||
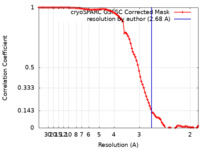
| Method | single particle reconstruction / cryo EM / Resolution: 2.68 Å | |||||||||
 Authors Authors | Li T / Hong J / Qu Q / Li D | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Molecular basis of the inositol deacylase PGAP1 involved in quality control of GPI-AP biogenesis. Authors: Jingjing Hong / Tingting Li / Yulin Chao / Yidan Xu / Zhini Zhu / Zixuan Zhou / Weijie Gu / Qianhui Qu / Dianfan Li /  Abstract: The secretion and quality control of glycosylphosphatidylinositol-anchored proteins (GPI-APs) necessitates post-attachment remodeling initiated by the evolutionarily conserved PGAP1, which deacylates ...The secretion and quality control of glycosylphosphatidylinositol-anchored proteins (GPI-APs) necessitates post-attachment remodeling initiated by the evolutionarily conserved PGAP1, which deacylates the inositol in nascent GPI-APs. Impairment of PGAP1 activity leads to developmental diseases in humans and fatality and infertility in animals. Here, we present three PGAP1 structures (2.66-2.84 Å), revealing its 10-transmembrane architecture and product-enzyme interaction details. PGAP1 holds GPI-AP acyl chains in an optimally organized, guitar-shaped cavity with apparent energetic penalties from hydrophobic-hydrophilic mismatches. However, abundant glycan-mediated interactions in the lumen counterbalance these repulsions, likely conferring substrate fidelity and preventing off-target hydrolysis of bulk membrane lipids. Structural and biochemical analyses uncover a serine hydrolase-type catalysis with atypical features and imply mechanisms for substrate entrance and product release involving a drawing compass movement of GPI-APs. Our findings advance the mechanistic understanding of GPI-AP remodeling. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36996.map.gz emd_36996.map.gz | 70.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36996-v30.xml emd-36996-v30.xml emd-36996.xml emd-36996.xml | 27.5 KB 27.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36996_fsc.xml emd_36996_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_36996.png emd_36996.png | 79.5 KB | ||
| Filedesc metadata |  emd-36996.cif.gz emd-36996.cif.gz | 8.8 KB | ||
| Others |  emd_36996_additional_1.map.gz emd_36996_additional_1.map.gz emd_36996_half_map_1.map.gz emd_36996_half_map_1.map.gz emd_36996_half_map_2.map.gz emd_36996_half_map_2.map.gz | 37.9 MB 69.8 MB 69.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36996 http://ftp.pdbj.org/pub/emdb/structures/EMD-36996 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36996 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36996 | HTTPS FTP |
-Related structure data
| Related structure data |  8k9rMC  8k9qC  8k9tC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36996.map.gz / Format: CCP4 / Size: 75.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36996.map.gz / Format: CCP4 / Size: 75.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.932 Å | ||||||||||||||||||||||||||||||||||||
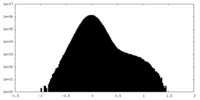
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_36996_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
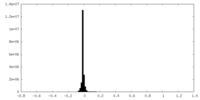
| Density Histograms |
-Half map: #2
| File | emd_36996_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
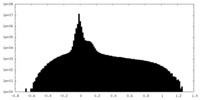
| Density Histograms |
-Half map: #1
| File | emd_36996_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
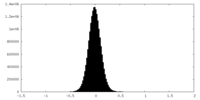
| Density Histograms |
- Sample components
Sample components
+Entire : Complex of the GPI inositol-deacylase with a chimera GPI-anchored...
+Supramolecule #1: Complex of the GPI inositol-deacylase with a chimera GPI-anchored...
+Macromolecule #1: GPI inositol-deacylase,MCherry protein
+Macromolecule #2: Green fluorescent protein,Complement decay-accelerating factor
+Macromolecule #3: alpha-D-mannopyranose
+Macromolecule #4: 2-azanylethyl [(2~{S},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-...
+Macromolecule #5: 2-amino-2-deoxy-alpha-D-glucopyranose
+Macromolecule #6: [(2~{R})-1-octadecoxy-3-[oxidanyl-[(2~{R},3~{R},5~{S},6~{R})-2,3,...
+Macromolecule #7: PALMITIC ACID
+Macromolecule #8: CHOLESTEROL
+Macromolecule #9: 2-azanylethyl [(2R,3S,4S,5S,6S)-3,4,5,6-tetrakis(oxidanyl)oxan-2-...
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY | ||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)