+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | rat megalin wingB | |||||||||
 Map data Map data | postprocess map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | endocytosis receptor / ENDOCYTOSIS | |||||||||
| Function / homology |  Function and homology information Function and homology informationchemoattraction of axon / diol metabolic process / positive regulation of oligodendrocyte progenitor proliferation / pulmonary artery morphogenesis / secondary heart field specification / folate import across plasma membrane / ventricular compact myocardium morphogenesis / response to leptin / metal ion transport / vitamin D metabolic process ...chemoattraction of axon / diol metabolic process / positive regulation of oligodendrocyte progenitor proliferation / pulmonary artery morphogenesis / secondary heart field specification / folate import across plasma membrane / ventricular compact myocardium morphogenesis / response to leptin / metal ion transport / vitamin D metabolic process / cranial skeletal system development / neuron projection arborization / coronary artery morphogenesis / outflow tract septum morphogenesis / aorta development / ventricular septum development / forebrain development / vagina development / positive regulation of endocytosis / endocytic vesicle / negative regulation of BMP signaling pathway / axonal growth cone / clathrin-coated pit / receptor-mediated endocytosis / endosome lumen / phosphatidylinositol 3-kinase/protein kinase B signal transduction / neural tube closure / kidney development / brush border membrane / sensory perception of sound / SH3 domain binding / male gonad development / protein transport / protein-folding chaperone binding / gene expression / cell population proliferation / receptor complex / external side of plasma membrane / calcium ion binding / dendrite / negative regulation of apoptotic process / endoplasmic reticulum / Golgi apparatus Similarity search - Function | |||||||||
| Biological species |  | |||||||||
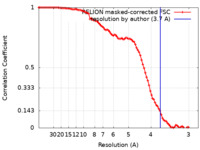
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Goto S / Tsutsumi A / Lee Y / Hosojima M / Kabasawa H / Komochi K / Yun-san L / Nagatoshi S / Tsumoto K / Nishizawa T ...Goto S / Tsutsumi A / Lee Y / Hosojima M / Kabasawa H / Komochi K / Yun-san L / Nagatoshi S / Tsumoto K / Nishizawa T / Kikkawa M / Saito A | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Cryo-EM structures elucidate the multiligand receptor nature of megalin. Authors: Sawako Goto / Akihisa Tsutsumi / Yongchan Lee / Michihiro Hosojima / Hideyuki Kabasawa / Koichi Komochi / Satoru Nagatoishi / Kazuya Takemoto / Kouhei Tsumoto / Tomohiro Nishizawa / Masahide ...Authors: Sawako Goto / Akihisa Tsutsumi / Yongchan Lee / Michihiro Hosojima / Hideyuki Kabasawa / Koichi Komochi / Satoru Nagatoishi / Kazuya Takemoto / Kouhei Tsumoto / Tomohiro Nishizawa / Masahide Kikkawa / Akihiko Saito /  Abstract: Megalin (low-density lipoprotein receptor-related protein 2) is a giant glycoprotein of about 600 kDa, mediating the endocytosis of more than 60 ligands, including those of proteins, peptides, and ...Megalin (low-density lipoprotein receptor-related protein 2) is a giant glycoprotein of about 600 kDa, mediating the endocytosis of more than 60 ligands, including those of proteins, peptides, and drug compounds [S. Goto, M. Hosojima, H. Kabasawa, A. Saito, , 106393 (2023)]. It is expressed predominantly in renal proximal tubule epithelial cells, as well as in the brain, lungs, eyes, inner ear, thyroid gland, and placenta. Megalin is also known to mediate the endocytosis of toxic compounds, particularly those that cause renal and hearing disorders [Y. Hori , , 1783-1791 (2017)]. Genetic megalin deficiency causes Donnai-Barrow syndrome/facio-oculo-acoustico-renal syndrome in humans. However, it is not known how megalin interacts with such a wide variety of ligands and plays pathological roles in various organs. In this study, we elucidated the dimeric architecture of megalin, purified from rat kidneys, using cryoelectron microscopy. The maps revealed the densities of endogenous ligands bound to various regions throughout the dimer, elucidating the multiligand receptor nature of megalin. We also determined the structure of megalin in complex with receptor-associated protein, a molecular chaperone for megalin. The results will facilitate further studies on the pathophysiology of megalin-dependent multiligand endocytic pathways in multiple organs and will also be useful for the development of megalin-targeted drugs for renal and hearing disorders, Alzheimer's disease [B. V. Zlokovic , , 4229-4234 (1996)], and other illnesses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36696.map.gz emd_36696.map.gz | 4.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36696-v30.xml emd-36696-v30.xml emd-36696.xml emd-36696.xml | 23.3 KB 23.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36696_fsc.xml emd_36696_fsc.xml | 9.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_36696.png emd_36696.png | 188.7 KB | ||
| Masks |  emd_36696_msk_1.map emd_36696_msk_1.map | 67 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-36696.cif.gz emd-36696.cif.gz | 9.1 KB | ||
| Others |  emd_36696_half_map_1.map.gz emd_36696_half_map_1.map.gz emd_36696_half_map_2.map.gz emd_36696_half_map_2.map.gz | 39.1 MB 39.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36696 http://ftp.pdbj.org/pub/emdb/structures/EMD-36696 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36696 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36696 | HTTPS FTP |
-Related structure data
| Related structure data |  8jxcMC  8jutC  8juuC  8jx8C  8jx9C  8jxaC  8jxbC  8jxdC  8jxeC  8jxfC  8jxgC  8jxhC  8jxiC  8jxjC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36696.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36696.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | postprocess map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.411 Å | ||||||||||||||||||||||||||||||||||||
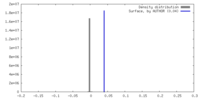
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_36696_msk_1.map emd_36696_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
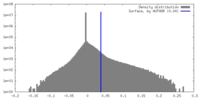
| Density Histograms |
-Half map: halfmap2
| File | emd_36696_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
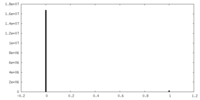
| Density Histograms |
-Half map: halfmap1
| File | emd_36696_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
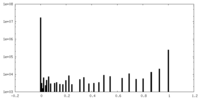
| Density Histograms |
- Sample components
Sample components
-Entire : megalin dimer at pH7.4
| Entire | Name: megalin dimer at pH7.4 |
|---|---|
| Components |
|
-Supramolecule #1: megalin dimer at pH7.4
| Supramolecule | Name: megalin dimer at pH7.4 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.2 MDa |
-Macromolecule #1: LDL receptor related protein 2
| Macromolecule | Name: LDL receptor related protein 2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 519.871438 KDa |
| Sequence | String: MERGAAAAAW MLLLAIAACL EPVSSQECGS GNFRCDNGYC IPASWRCDGT RDCLDDTDEI GCPPRSCESG LFLCPAEGTC IPSSWVCDE DKDCSDGADE QQNCAGTTCS AQQMTCSNGQ CIPSEYRCDH VSDCPDGSDE RNCHYPTCDQ LTCANGACYN T SQRCDQKV ...String: MERGAAAAAW MLLLAIAACL EPVSSQECGS GNFRCDNGYC IPASWRCDGT RDCLDDTDEI GCPPRSCESG LFLCPAEGTC IPSSWVCDE DKDCSDGADE QQNCAGTTCS AQQMTCSNGQ CIPSEYRCDH VSDCPDGSDE RNCHYPTCDQ LTCANGACYN T SQRCDQKV DCRDSSDEAN CTTLCSQKEF ECGSGECILR AYVCDHDNDC EDNSDERNCN YDTCGGHQFT CSNGQCINQN WV CDGDDDC QDSGDEDGCE SNQSHHRCYP REWACPGSGR CISIDKVCDG VPDCPEGDDE NNVTSGRTCG MGVCSVLNCE YQC HQTPFG GECFCPPGHI INSNDSRTCI DFDDCQIWGI CDQKCENRQG RHQCLCEEGY ILERGQHCKS SDSFSAASVI FSNG RDLLV GDLHGRNFRI LAESKNRGMV MGVDFHYQKH RVFWTDPMQE KVFSTDINGL NTQEILNVSV DTPENLAVDW INNKL YLVE TKVNRIDVVN LEGNQRVTLI TENLGHPRGI ALDPTVGYLF FSDWGSLSGQ PKVERAFMDG SNRKDLVTTK VGWPAG ITL DLVSKRVYWV DSRYDYIETV TYDGIQRKTV ARGGSLVPHP FGISLFEEHV FFTDWTKMAV MKASKFTETN PQVYHQS SL RPHGVTVYHA LRQPNATNPC GSNNGGCAQV CVLSHRTDNG GLGYRCKCEF GFELDDDEHR CVAVKNFLLF SSKTAVRG I PFTLSTQEDV MVPVTGSPSF FVGIDFDAQH STVFYSDLSK DIIYKQKIDG TGKEVITANR LESVECLTFD WISRNLYWT DGGLKSVTVL RLADKSRRQI ISNLNNPRSI VVHPTAGYMF LSDWFRPAKI MRAWSDGSHL MPIVNTSLGW PNGLAIDWSA SRLYWVDAF FDKIEHSTLD GLDRKRLGHV DQMTHPFGLT VFKDNVFITD WRLGAIIRVR KSDGGDMTVI RRGISSVMHV K AYDADLQT GSNYCSQTTH ANGDCSHFCF PVPNFQRVCG CPYGMKLQRD QMTCEGDPAR EPPTQQCGSL SFPCNNGKCV PS FFRCDGV DDCHDNSDEH QCGVFNNTCS PSAFACVRGG QCIPGQWHCD RQNDCLDGSD EQNCPTHATS STCPSTSFTC DNH VCIPKD WVCDTDNDCS DGSDEKNCQA SGTCQPTQFR CPDHRCISPL YVCDGDKDCA DGSDEAGCVL NCTSAQFKCA DGSS CINSR YRCDGVYDCR DNSDEAGCPT RPPGMCHLDE FQCQGDGTCI PNTWECDGHP DCIHGSDEHT GCVPKTCSPT HFLCD NGNC IYKAWICDGD NDCRDMSDEK DCPTQPFHCP STQWQCPGYS TCINLSALCD GVFDCPNGTD ESPLCNQDSC SHFNGG CTH QCMQGPFGAT CLCPLGYQLA NDTKTCEDIN ECDIPGFCSQ HCVNMRGSFR CACDPEYTLE SDGRTCKVTG SENPLLV VA SRDKIIVDNI TAHTHNLYSL VQDVSFVVAL DFDSVTGRVF WSDLLQGKTW SVFQNGTDKR VVHDSGLSVT EMIAVDWI G RNLYWTDYAL ETIEVSKIDG SHRTVLISKN VTKPRGLALD PRMGDNVMFW SDWGHHPRIE RASMDGTMRT VIVQEKIYW PCGLSIDYPN RLIYFMDAYL DYIEFCDYDG HNRRQVIASD LVLHHPHALT LFEDFVYWTD RGTRQVMQAN KWHGGNQSVV MYSVHQPLG ITAIHPSRQP PSRNPCASAS CSHLCLLSAQ APRHYSCACP SGWNLSDDSV NCVRGDQPFL MSVRDNIIFG I SLDPEVKS NDAMVPISGI QHGYDVEFDD SEQFIYWVEN PGEIHRVKTD GSNRTVFAPL SLLGSSLGLA LDWVSRNIYY TT PASRSIE VLTLKGDTRY GKTLIANDGT PLGVGFPVGI AVDPARGKLY WSDHGTDSGV PAKIASANMD GTSLKILFTG NLQ HLEVVT LDIQEQKLYW AVTSRGVIER GNVDGTERMI LVHHLAHPWG LVVYGSFLYY SDEQYEVIER VDKSSGNNKV VLRD NVPYL RGLRVYHRRN AADSSNGCSN NPNACQQICL PVPGGMFSCA CASGFKLSPD GRSCSPYNSF MVVSMLPAVR GFSLE LSDH SEAMVPVAGQ GRNVLHADVD VANGFIYWCD FSSSVRSSNG IRRIKPDGSN FTNVVTYGIG ANGIRGVALD WAAGNL YFT NAFVYETLIE VLRINTTYRR VLLKVSVDMP RHIIVDPKHR YLFWADYGQK PKIERSFLDC TNRTVLVSEG IVTPRGL AM DHDTGYIYWV DDSLDLIARI HLDGGESQVV RYGSRYPTPY GITVFGESII WVDRNLKKVF QASKQPGNTD PPVVIRDK I NLLRDVTIFD EHAQPLSPAE LNNNPCLQSN GGCSHFCFAL PELPTPRCGC AFGTLGNDGK SCATSQEDFL IYSLNNSLR SLHFDPRDHS LPFQVISVAG TAIALDYDRR NNRIFFTQKL NSLRGQISYV SLYSGSSSPT VLLSNIGVTD GIAFDWINRR IYYSDFSNQ TINSMAEDGS NRAVIARVSK PRAIVLDPCR GYMYWTDWGT NAKIERATLG GNFRVPIVNT SLVWPNGLAL D LETDLLYW ADASLQKIER STLTGTNREV VVSTAFHSFG LTVYGQYIYW TDLYTRKIYR ANKYDGSDLV AMTTRLPTQP SG ISTVVKT QRQQCSNPCD QFNGGCSHIC APGPNGAECQ CPHEGNWYLA NDNKYCVVDT GTRCNQLQFT CLNGHCINQD WKC DNDNDC GDGSDELPTV CAFHTCRSTA FTCGNGRCVP YHYRCDYYND CGDNSDEAGC LFRNCNSTTE FTCSNGRCIP LSYV CNGIN NCHDNDTSDE KNCPPHTCPP DFTKCQTTNI CVPRAFLCDG DNDCGDGSDE NPIYCASHTC RSNEFQCLSP QRCIP SYWF CDGEADCADG SDEPDTCGHS VNTCRASQFQ CDNGRCISGN WVCDGDNDCG DMSDEDQRHH CELQNCSSTQ FTCVNS RPP NRRCIPQYWV CDGDADCSDA LDELQNCTMR TCSAGEFSCA NGRCVRQSFR CDRRNDCGDY SDERGCSYPP CHANQFT CQ NGRCIPRFFV CDEDNDCGDG SDEQEHLCHT PEPTCPLHQF RCDNGHCIEM GRVCNHVDDC SDNSDEKGCG INECLDSS I SRCDHNCTDT ITSFYCSCLP GYKLMSDKRS CVDIDECKES PQLCSQKCEN VVGSYICKCA PGYIREPDGK SCRQNSNIE PYLIFSNRYY IRNLTTDGSS YSLILQGLGN VVALDFDRVE KRLYWIDAEK QIIERMFLNK TNRETIINHR LRRAESLAVD WVSRKLYWL DAILDCLFVS DLEGRHRKMI AQHCVDANNT FCFEHPRGIV LHPQRGHVYW ADWGVHAYIG RIGMDGTNKS V IISTKIEW PNAITIDYTN DLLYWADAHL GYIEFSDLEG HHRHTVYDGS LPHPFALTIF EDTVFWTDWN TRTVEKGNKY DG SGRVVLV NTTHKPFDIH VYHPYRQPIM SNPCGTNNGG CSHLCLIKAG GRGFTCACPD DFQTVQLRDR TLCMPMCSST QFL CGNNEK CIPIWWKCDG QKDCSDGSDE PDLCPHRFCR LGQFQCRDGN CTSPQALCNA RQDCADGSDE DRVLCEHHRC ESNE WQCAN KRCIPQSWQC DSVNDCLDNS DEDTSHCASR TCRPGQFKCN NGRCIPQSWK CDVDNDCGDY SDEPIDECTT AAYNC DNHT EFSCKTNYRC IPQWAVCNGF DDCRDNSDEQ GCESVPCHPS GDFRCANHHC IPLRWKCDGT DDCGDNSDEE NCVPRE CSE SEFRCADQQC IPSRWVCDQE NDCGDNSDER DCEMKTCHPE HFQCTSGHCV PKALACDGRA DCLDASDESA CPTRFPN GT YCPAAMFECK NHVCIQSFWI CDGENDCVDG SDEEIHLCFN IPCESPQRFR CDNSRCVYGH QLCNGVDDCG DGSDEKEE H CRKPTHKPCT DTEYKCSNGN CISQHYVCDN VNDCGDLSDE TGCNLGDNRT CAENICEQNC TQLSSGGFIC SCRPGFKPS TLDKNSCQDI NECEEFGICP QSCRNSKGSY ECFCVDGFKS MSTHYGERCA ADGSPPLLLL PENVRIRKYN TSSEKFSEYL EEEEHIQTI DYDWDPEHIG LSVVYYTVLA QGSQFGAIKR AYIPNFESGS NNPIREVDLG LKYLMQPDGL AVDWVGRHIY W SDAKSQRI EVATLDGRYR KWLITTQLDQ PAAIAVNPKL GLMFWTDQGK QPKIESAWMN GEHRSVLVSE NLGWPNGLSI DY LNDDRVY WSDSKEDVIE AIKYDGTDRR LIINEAMKPF SLDIFEDKLY WVAKEKGEVW RQNKFGKENK EKVLVVNPWL TQV RIFHQL RYNQSVSNPC KQVCSHLCLL RPGGYSCACP QGSDFVTGST VQCDAASELP VTMPPPCRCM HGGNCYFDEN ELPK CKCSS GYSGEYCEVG LSRGIPPGTT MAVLLTFVIV IIVGALVLVG LFHYRKTGSL LPTLPKLPSL SSLAKPSENG NGVTF RSGA DVNMDIGVSP FGPETIIDRS MAMNEHFVME VGKQPVIFEN PMYAAKDNTS KVALAVQGPS TGAQVTVPEN VENQNY GRP IDPSEIVPEP KPASPGADEI QGKKWNIFKR KPKQTTNFEN PIYAEMDSEV KDAVAVAPPP SPSLPAKASK RNLTPGY TA TEDTFKDTAN LVKEDSDV UniProtKB: Low-density lipoprotein receptor-related protein 2 |
-Macromolecule #2: unclear peptide
| Macromolecule | Name: unclear peptide / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 528.644 Da |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
-Macromolecule #3: unclear peptide
| Macromolecule | Name: unclear peptide / type: protein_or_peptide / ID: 3 Details: Authors could not experimentally confirm this sequence, but it should be a part of molecule 1. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 472.537 Da |
| Sequence | String: (UNK)N(UNK)(UNK)(UNK) |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 5 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #7: 2-acetamido-2-deoxy-alpha-D-galactopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-alpha-D-galactopyranose / type: ligand / ID: 7 / Number of copies: 8 / Formula: A2G |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-A2G: |
-Macromolecule #8: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 8 / Number of copies: 17 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.4 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-8jxc: |
 Movie
Movie Controller
Controller






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)