+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | rat megalin RAP complex | |||||||||
 Map data Map data | postprocess consensus map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | endocytosis receptor / ENDOCYTOSIS | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of receptor-mediated endocytosis / negative regulation of very-low-density lipoprotein particle clearance / chemoattraction of axon / diol metabolic process / positive regulation of oligodendrocyte progenitor proliferation / pulmonary artery morphogenesis / secondary heart field specification / rough endoplasmic reticulum lumen / lipase binding / folate import across plasma membrane ...regulation of receptor-mediated endocytosis / negative regulation of very-low-density lipoprotein particle clearance / chemoattraction of axon / diol metabolic process / positive regulation of oligodendrocyte progenitor proliferation / pulmonary artery morphogenesis / secondary heart field specification / rough endoplasmic reticulum lumen / lipase binding / folate import across plasma membrane / receptor antagonist activity / amyloid-beta clearance by transcytosis / negative regulation of amyloid-beta clearance / ventricular compact myocardium morphogenesis / response to leptin / metal ion transport / vitamin D metabolic process / very-low-density lipoprotein particle receptor binding / cranial skeletal system development / neuron projection arborization / coronary artery morphogenesis / cis-Golgi network / negative regulation of receptor internalization / outflow tract septum morphogenesis / ventricular septum development / aorta development / low-density lipoprotein particle receptor binding / endoplasmic reticulum-Golgi intermediate compartment / forebrain development / vagina development / positive regulation of endocytosis / negative regulation of BMP signaling pathway / endocytic vesicle / axonal growth cone / clathrin-coated pit / endomembrane system / receptor-mediated endocytosis / endosome lumen / phosphatidylinositol 3-kinase/protein kinase B signal transduction / kidney development / neural tube closure / brush border membrane / sensory perception of sound / SH3 domain binding / Golgi lumen / male gonad development / heparin binding / protein transport / protein-folding chaperone binding / gene expression / vesicle / receptor complex / cell population proliferation / endosome / receptor ligand activity / external side of plasma membrane / calcium ion binding / dendrite / negative regulation of apoptotic process / cell surface / endoplasmic reticulum / Golgi apparatus / signal transduction / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
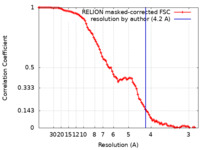
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Goto S / Tsutsumi A / Lee Y / Hosojima M / Kabasawa H / Komochi K / Yun-san L / Nagatoshi S / Tsumoto K / Nishizawa T ...Goto S / Tsutsumi A / Lee Y / Hosojima M / Kabasawa H / Komochi K / Yun-san L / Nagatoshi S / Tsumoto K / Nishizawa T / Kikkawa M / Saito A | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Cryo-EM structures elucidate the multiligand receptor nature of megalin. Authors: Sawako Goto / Akihisa Tsutsumi / Yongchan Lee / Michihiro Hosojima / Hideyuki Kabasawa / Koichi Komochi / Satoru Nagatoishi / Kazuya Takemoto / Kouhei Tsumoto / Tomohiro Nishizawa / Masahide ...Authors: Sawako Goto / Akihisa Tsutsumi / Yongchan Lee / Michihiro Hosojima / Hideyuki Kabasawa / Koichi Komochi / Satoru Nagatoishi / Kazuya Takemoto / Kouhei Tsumoto / Tomohiro Nishizawa / Masahide Kikkawa / Akihiko Saito /  Abstract: Megalin (low-density lipoprotein receptor-related protein 2) is a giant glycoprotein of about 600 kDa, mediating the endocytosis of more than 60 ligands, including those of proteins, peptides, and ...Megalin (low-density lipoprotein receptor-related protein 2) is a giant glycoprotein of about 600 kDa, mediating the endocytosis of more than 60 ligands, including those of proteins, peptides, and drug compounds [S. Goto, M. Hosojima, H. Kabasawa, A. Saito, , 106393 (2023)]. It is expressed predominantly in renal proximal tubule epithelial cells, as well as in the brain, lungs, eyes, inner ear, thyroid gland, and placenta. Megalin is also known to mediate the endocytosis of toxic compounds, particularly those that cause renal and hearing disorders [Y. Hori , , 1783-1791 (2017)]. Genetic megalin deficiency causes Donnai-Barrow syndrome/facio-oculo-acoustico-renal syndrome in humans. However, it is not known how megalin interacts with such a wide variety of ligands and plays pathological roles in various organs. In this study, we elucidated the dimeric architecture of megalin, purified from rat kidneys, using cryoelectron microscopy. The maps revealed the densities of endogenous ligands bound to various regions throughout the dimer, elucidating the multiligand receptor nature of megalin. We also determined the structure of megalin in complex with receptor-associated protein, a molecular chaperone for megalin. The results will facilitate further studies on the pathophysiology of megalin-dependent multiligand endocytic pathways in multiple organs and will also be useful for the development of megalin-targeted drugs for renal and hearing disorders, Alzheimer's disease [B. V. Zlokovic , , 4229-4234 (1996)], and other illnesses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36663.map.gz emd_36663.map.gz | 42.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36663-v30.xml emd-36663-v30.xml emd-36663.xml emd-36663.xml | 30.7 KB 30.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36663_fsc.xml emd_36663_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_36663.png emd_36663.png | 169.4 KB | ||
| Masks |  emd_36663_msk_1.map emd_36663_msk_1.map | 67 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-36663.cif.gz emd-36663.cif.gz | 9.8 KB | ||
| Others |  emd_36663_additional_1.map.gz emd_36663_additional_1.map.gz emd_36663_half_map_1.map.gz emd_36663_half_map_1.map.gz emd_36663_half_map_2.map.gz emd_36663_half_map_2.map.gz | 28.6 MB 52.2 MB 52.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36663 http://ftp.pdbj.org/pub/emdb/structures/EMD-36663 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36663 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36663 | HTTPS FTP |
-Related structure data
| Related structure data |  8jutMC  8juuC  8jx8C  8jx9C  8jxaC  8jxbC  8jxcC  8jxdC  8jxeC  8jxfC  8jxgC  8jxhC  8jxiC  8jxjC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36663.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36663.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | postprocess consensus map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.411 Å | ||||||||||||||||||||||||||||||||||||
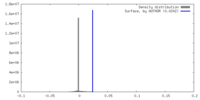
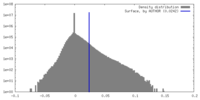
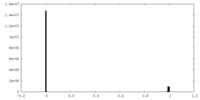
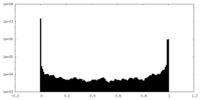
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_36663_msk_1.map emd_36663_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
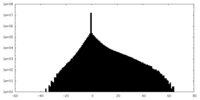
| Density Histograms |
-Additional map: composite map
| File | emd_36663_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | composite map | ||||||||||||
| Projections & Slices |
| ||||||||||||
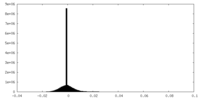
| Density Histograms |
-Half map: halfmap1
| File | emd_36663_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
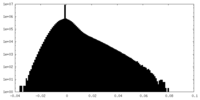
| Density Histograms |
-Half map: halfmap2
| File | emd_36663_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
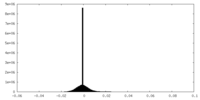
| Density Histograms |
- Sample components
Sample components
+Entire : megalin-RAP complex
+Supramolecule #1: megalin-RAP complex
+Macromolecule #1: LDL receptor related protein 2
+Macromolecule #2: Alpha-2-macroglobulin receptor-associated protein
+Macromolecule #3: unclear peptide
+Macromolecule #4: unclear peptide
+Macromolecule #5: unclear peptide
+Macromolecule #6: unclear peptide
+Macromolecule #7: unclear peptide
+Macromolecule #8: unclear peptide
+Macromolecule #17: 2-acetamido-2-deoxy-beta-D-glucopyranose
+Macromolecule #18: 2-acetamido-2-deoxy-alpha-D-galactopyranose
+Macromolecule #19: CALCIUM ION
+Macromolecule #20: NICKEL (II) ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)