+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Fiber I and fiber-tail-adaptor of phage GP4 | |||||||||||||||
 Map data Map data | portal-vertex of phage GP4 | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Complex / VIRAL PROTEIN | |||||||||||||||
| Function / homology | Virion-associated phage protein / Virion-associated phage protein Function and homology information Function and homology information | |||||||||||||||
| Biological species |  Ralstonia phage GP4 (virus) Ralstonia phage GP4 (virus) | |||||||||||||||
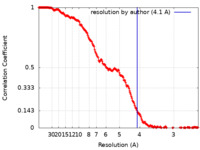
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||||||||
 Authors Authors | Liu H / Chen W | |||||||||||||||
| Funding support |  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2023 Journal: J Mol Biol / Year: 2023Title: Asymmetric Structure of Podophage GP4 Reveals a Novel Architecture of Three Types of Tail Fibers. Authors: Jing Zheng / Wenyuan Chen / Hao Xiao / Fan Yang / Jingdong Song / Lingpeng Cheng / Hongrong Liu /  Abstract: Bacteriophage tail fibers (or called tail spikes) play a critical role in the early stage of infection by binding to the bacterial surface. Podophages with known structures usually possess one or two ...Bacteriophage tail fibers (or called tail spikes) play a critical role in the early stage of infection by binding to the bacterial surface. Podophages with known structures usually possess one or two types of fibers. Here, we resolved an asymmetric structure of the podophage GP4 to near-atomic resolution by cryo-EM. Our structure revealed a symmetry-mismatch relationship between the components of the GP4 tail with previously unseen topologies. In detail, two dodecameric adaptors (adaptors I and II), a hexameric nozzle, and a tail needle form a conserved tail body connected to a dodecameric portal occupying a unique vertex of the icosahedral head. However, five chain-like extended fibers (fiber I) and five tulip-like short fibers (fiber II) are anchored to a 15-fold symmetric fiber-tail adaptor, encircling the adaptor I, and six bamboo-like trimeric fibers (fiber III) are connected to the nozzle. Five fibers I, each composed of five dimers of the protein gp80 linked by an elongated rope protein, are attached to the five edges of the tail vertex of the icosahedral head. In this study, we identified a new structure of the podophage with three types of tail fibers, and such phages with different types of fibers may have a broad host range and/or infect host cells with considerably high efficiency, providing evolutionary advantages in harsh environments. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36462.map.gz emd_36462.map.gz | 580.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36462-v30.xml emd-36462-v30.xml emd-36462.xml emd-36462.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36462_fsc.xml emd_36462_fsc.xml | 21.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_36462.png emd_36462.png | 174.6 KB | ||
| Filedesc metadata |  emd-36462.cif.gz emd-36462.cif.gz | 5.1 KB | ||
| Others |  emd_36462_half_map_1.map.gz emd_36462_half_map_1.map.gz emd_36462_half_map_2.map.gz emd_36462_half_map_2.map.gz | 582.1 MB 582.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36462 http://ftp.pdbj.org/pub/emdb/structures/EMD-36462 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36462 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36462 | HTTPS FTP |
-Validation report
| Summary document |  emd_36462_validation.pdf.gz emd_36462_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36462_full_validation.pdf.gz emd_36462_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_36462_validation.xml.gz emd_36462_validation.xml.gz | 27.6 KB | Display | |
| Data in CIF |  emd_36462_validation.cif.gz emd_36462_validation.cif.gz | 37.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36462 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36462 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36462 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36462 | HTTPS FTP |
-Related structure data
| Related structure data |  8jouMC  8jovC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36462.map.gz / Format: CCP4 / Size: 641.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36462.map.gz / Format: CCP4 / Size: 641.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | portal-vertex of phage GP4 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.27 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map2
| File | emd_36462_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map1
| File | emd_36462_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ralstonia phage GP4
| Entire | Name:  Ralstonia phage GP4 (virus) Ralstonia phage GP4 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Ralstonia phage GP4
| Supramolecule | Name: Ralstonia phage GP4 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2282904 / Sci species name: Ralstonia phage GP4 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Virion-associated phage protein
| Macromolecule | Name: Virion-associated phage protein / type: protein_or_peptide / ID: 1 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Ralstonia phage GP4 (virus) Ralstonia phage GP4 (virus) |
| Molecular weight | Theoretical: 14.360909 KDa |
| Sequence | String: MAAASTYTEN NILNALLRGV AFPLPAKTYV SLHTGDPGVG AGANEVSLSN WPAYVRREAE QGGAIGSGWT PAASGQTSNV NQLTYPANN GVAAVTVTHY AVFDAPTGGN LLFKAPLTVA RTLQVGDVFV FDVGSLTAQA S UniProtKB: Virion-associated phage protein |
-Macromolecule #2: rope protein of phage GP4
| Macromolecule | Name: rope protein of phage GP4 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Ralstonia phage GP4 (virus) Ralstonia phage GP4 (virus) |
| Molecular weight | Theoretical: 10.230603 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
-Macromolecule #3: Virion-associated phage protein
| Macromolecule | Name: Virion-associated phage protein / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Ralstonia phage GP4 (virus) Ralstonia phage GP4 (virus) |
| Molecular weight | Theoretical: 9.908155 KDa |
| Sequence | String: MLGIVQKTAT EQLDYDIDFA RWMPDGDVLQ SAGVVITPDD GTLTSPAYEI DGTVVKVWLA GGTAGASYNV DVTVATAAGR IKETCFKTR VRSC UniProtKB: Virion-associated phage protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.8000000000000003 µm / Nominal defocus min: 0.1 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)