Yorodumi
Yorodumi+ Open data
Open data
 Loading...
Loading...
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - State Spb4 | |||||||||||||||
 Map data Map data | local resolution filtered map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Ribosome / Ribosome biogenesis / pre-60S ribosome | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationNoc1p-Noc2p complex / regulation of cytoplasmic translational initiation / Noc2p-Noc3p complex / rRNA (uridine-2'-O-)-methyltransferase activity / rRNA (guanine) methyltransferase activity / dolichyl-diphosphooligosaccharide-protein glycotransferase / rRNA (cytosine-C5-)-methyltransferase activity / dolichyl-diphosphooligosaccharide-protein glycotransferase activity / intracellular mRNA localization / oligosaccharyltransferase complex ...Noc1p-Noc2p complex / regulation of cytoplasmic translational initiation / Noc2p-Noc3p complex / rRNA (uridine-2'-O-)-methyltransferase activity / rRNA (guanine) methyltransferase activity / dolichyl-diphosphooligosaccharide-protein glycotransferase / rRNA (cytosine-C5-)-methyltransferase activity / dolichyl-diphosphooligosaccharide-protein glycotransferase activity / intracellular mRNA localization / oligosaccharyltransferase complex / preribosome / PeBoW complex / : / maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / rRNA base methylation / ribosomal large subunit binding / preribosome, large subunit precursor / nuclear-transcribed mRNA catabolic process / DNA replication initiation / ribosomal large subunit export from nucleus / protein kinase activator activity / mRNA transport / ribonucleoprotein complex binding / ribosomal subunit export from nucleus / protein-RNA complex assembly / maturation of LSU-rRNA / translation initiation factor activity / nuclear periphery / post-translational protein modification / cellular response to amino acid starvation / assembly of large subunit precursor of preribosome / cytosolic ribosome assembly / ribosome assembly / ribosomal large subunit biogenesis / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / rRNA processing / protein transport / large ribosomal subunit / ribosome biogenesis / ribosomal large subunit assembly / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / RNA helicase activity / negative regulation of translation / rRNA binding / structural constituent of ribosome / RNA helicase / ribosome / translation / ribonucleoprotein complex / mRNA binding / chromatin binding / GTP binding / nucleolus / ATP hydrolysis activity / RNA binding / zinc ion binding / nucleoplasm / ATP binding / metal ion binding / cytoplasm Similarity search - Function : / ATP-dependent rRNA helicase SPB4-like, C-terminal tail / 60S ribosomal subunit assembly/export protein Loc1 / : / Methyltr_RsmF/B-like, ferredoxin-like domain / Nucleolar complex protein 2 / Nucleolar complex-associated protein 3, N-terminal / Nucleolar complex-associated protein 3 / Noc2p family / Nucleolar complex-associated protein ...: / ATP-dependent rRNA helicase SPB4-like, C-terminal tail / 60S ribosomal subunit assembly/export protein Loc1 / : / Methyltr_RsmF/B-like, ferredoxin-like domain / Nucleolar complex protein 2 / Nucleolar complex-associated protein 3, N-terminal / Nucleolar complex-associated protein 3 / Noc2p family / Nucleolar complex-associated protein / Gcn1, N-terminal / : / : / : / Generalcontrol nonderepressible 1 (Gcn1) N-terminal / Stalled ribosome sensor GCN1-like, HEAT repeats / Fungal GCN1-like, HEAT repeats / Stalled ribosome sensor GCN1 N-terminal / Stalled ribosome sensor GCN1-like, HEAT repeats / WD repeat WDR12/Ytm1 / Ribosome biogenesis factor, NIP7 / RNA (C5-cytosine) methyltransferase, NOP2 / : / E-Z type HEAT repeats / PBS lyase HEAT-like repeat / UPF0113, pre-PUA domain / UPF0113 PUA domain / NIP7/UPF0113, pre-PUA domain / UPF0113, PUA domain / Elongation factor 3/GNC1 HEAT repeat / EF3 N-terminal HEAT repeat / Centrosomal protein CEP104, TOG domain / Nop2p / Bacterial Fmu (Sun)/eukaryotic nucleolar NOL1/Nop2p, conserved site / NOL1/NOP2/sun family signature. / : / STT3/PglB/AglB core domain / Ribosomal RNA methyltransferase, SPB1-like, C-terminal / Ribosomal RNA methyltransferase Spb1, domain of unknown function DUF3381 / AdoMet-dependent rRNA methyltransferase SPB1-like / Spb1 C-terminal domain / Ribosomal RNA methyltransferase Spb1, DUF3381 / Cgr1-like / Cgr1 family / Eukaryotic rRNA processing / Eukaryotic rRNA processing protein EBP2 / : / Ribosomal RNA large subunit methyltransferase E / Parkin co-regulated protein / Ribosome biogenesis protein BRX1 / TOG domain / Parkin co-regulated protein / TOG / DDX18/Has1, DEAD-box helicase domain / Ribosome biogenesis protein Nop16 / Ribosome biogenesis protein Nop16 / Domain of unknown function DUF4217 / ATP-dependent rRNA helicase SPB4-like, C-terminal extension / DUF4217 / Domain of unknown function DUF2423 / YBL028C ribosome biogenesis factor, N-terminal domain / BOP1, N-terminal domain / WD repeat BOP1/Erb1 / BOP1NT (NUC169) domain / BOP1NT (NUC169) domain / : / Oligosaccharyl transferase, STT3 subunit / Oligosaccharyl transferase STT3, N-terminal / NLE / NLE (NUC135) domain / SAM-dependent methyltransferase RsmB/NOP2-type / RNA (C5-cytosine) methyltransferase / : / 16S rRNA methyltransferase RsmB/F / SAM-dependent MTase RsmB/NOP-type domain profile. / Guanine nucleotide-binding protein-like 3, N-terminal domain / GNL3L/Grn1 putative GTPase / CCAAT-binding factor / CBF/Mak21 family / Putative RNA-binding Domain in PseudoUridine synthase and Archaeosine transglycosylase / PUA domain / PUA domain superfamily / : / PUA domain profile. / Pescadillo / Pescadillo N-terminus / Ribosomal biogenesis NSA2 family / GTP-binding protein, orthogonal bundle domain superfamily / Ribosome assembly factor Mrt4 / : / NOG, C-terminal / Nucleolar GTP-binding protein 1 / NOGCT (NUC087) domain / Nucleolar GTP-binding protein 1, Rossman-fold domain / NOG1, N-terminal helical domain / Nucleolar GTP-binding protein 1 (NOG1) / NOG1 N-terminal helical domain / Circularly permuted (CP)-type guanine nucleotide-binding (G) domain / Circularly permuted (CP)-type guanine nucleotide-binding (G) domain profile. / Brix domain Similarity search - Domain/homology Large ribosomal subunit protein uL3 / Brix domain-containing protein / ATP-dependent RNA helicase / 60S ribosomal protein L26-like protein / ATP-dependent RNA helicase / Ribosomal protein L15 / 60S ribosomal protein L27 / Ribosome biogenesis protein NSA2 homolog / 60S ribosomal protein L6 / Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3 ...Large ribosomal subunit protein uL3 / Brix domain-containing protein / ATP-dependent RNA helicase / 60S ribosomal protein L26-like protein / ATP-dependent RNA helicase / Ribosomal protein L15 / 60S ribosomal protein L27 / Ribosome biogenesis protein NSA2 homolog / 60S ribosomal protein L6 / Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3 / 60S ribosomal protein l9-like protein / 60S ribosomal protein l21-like protein / Ribosomal protein L37 / Putative RNA-binding protein / 60S ribosomal protein L14-like protein / 60S ribosome subunit biogenesis protein NIP7 / Ribosome biogenesis protein RLP24 / Ribosomal protein / 60S ribosomal protein L25-like protein / Nucleolar complex-associated protein 3 / Large ribosomal subunit protein uL10 / Eukaryotic translation initiation factor 6 / 60S ribosomal protein L32-like protein / Uncharacterized protein / Nucleolar protein 16 / Nucleolar GTP-binding protein 1 / 60S ribosomal protein L13 / Ribosomal protein L18-like protein / Ribosomal protein L19 / 60S ribosomal protein L8 / 60S ribosomal protein L36 / 60S ribosomal protein L20 / Ribosome biogenesis protein ERB1 / 60S ribosomal protein l33-like protein / rRNA-processing protein / Putative NOC2 family protein / Putative 60S ribosomal protein / 60S ribosomal protein L22-like protein / 60S ribosomal protein l30-like protein / Putative GTP binding protein / Ribosome biogenesis protein YTM1 / 60S ribosomal protein L4-like protein / 60S ribosomal protein l7-like protein / Ribosomal protein l34-like protein / 60S ribosomal protein L38-like protein / rRNA processing protein / Uncharacterized protein / 60S ribosomal protein L12-like protein / 60S ribosomal subunit-like protein / Large ribosomal subunit protein uL22 / 60S ribosomal protein L16-like protein / Putative 60S ribosomal protein / DUF2423 domain-containing protein / Nucleolar protein 2 / Pescadillo homolog / 60S ribosomal protein l23-like protein / Large ribosomal subunit protein eL39 Similarity search - Component | |||||||||||||||
| Biological species |  Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) | |||||||||||||||
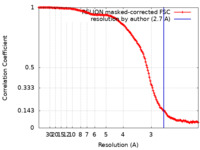
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||||||||
 Authors Authors | Lau B / Huang Z / Beckmann R / Hurt E / Cheng J | |||||||||||||||
| Funding support | 1 items
| |||||||||||||||
 Citation Citation |  Journal: EMBO Rep / Year: 2023 Journal: EMBO Rep / Year: 2023Title: Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis. Authors: Benjamin Lau / Zixuan Huang / Nikola Kellner / Shuangshuang Niu / Otto Berninghausen / Roland Beckmann / Ed Hurt / Jingdong Cheng /   Abstract: Ribosome biogenesis proceeds along a multifaceted pathway from the nucleolus to the cytoplasm that is extensively coupled to several quality control mechanisms. However, the mode by which 5S ...Ribosome biogenesis proceeds along a multifaceted pathway from the nucleolus to the cytoplasm that is extensively coupled to several quality control mechanisms. However, the mode by which 5S ribosomal RNA is incorporated into the developing pre-60S ribosome, which in humans links ribosome biogenesis to cell proliferation by surveillance by factors such as p53-MDM2, is poorly understood. Here, we report nine nucleolar pre-60S cryo-EM structures from Chaetomium thermophilum, one of which clarifies the mechanism of 5S RNP incorporation into the early pre-60S. Successive assembly states then represent how helicases Dbp10 and Spb4, and the Pumilio domain factor Puf6 act in series to surveil the gradual folding of the nearby 25S rRNA domain IV. Finally, the methyltransferase Spb1 methylates a universally conserved guanine nucleotide in the A-loop of the peptidyl transferase center, thereby licensing further maturation. Our findings provide insight into the hierarchical action of helicases in safeguarding rRNA tertiary structure folding and coupling to surveillance mechanisms that culminate in local RNA modification. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35289.map.gz emd_35289.map.gz | 162.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35289-v30.xml emd-35289-v30.xml emd-35289.xml emd-35289.xml | 98.5 KB 98.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35289_fsc.xml emd_35289_fsc.xml | 14.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_35289.png emd_35289.png | 161.5 KB | ||
| Filedesc metadata |  emd-35289.cif.gz emd-35289.cif.gz | 22.5 KB | ||
| Others |  emd_35289_additional_1.map.gz emd_35289_additional_1.map.gz emd_35289_half_map_1.map.gz emd_35289_half_map_1.map.gz emd_35289_half_map_2.map.gz emd_35289_half_map_2.map.gz | 254.4 MB 225.3 MB 225.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35289 http://ftp.pdbj.org/pub/emdb/structures/EMD-35289 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35289 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35289 | HTTPS FTP |
-Related structure data
| Related structure data |  8i9zMC  8i9pC  8i9rC  8i9tC  8i9vC  8i9wC  8i9xC  8i9yC  8ia0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35289.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35289.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local resolution filtered map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.059 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: DeepEMhancer filtered map
| File | emd_35289_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer filtered map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_35289_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_35289_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : pre-60S ribosome
| Entire | Name: pre-60S ribosome |
|---|---|
| Components |
|
+Supramolecule #1: pre-60S ribosome
| Supramolecule | Name: pre-60S ribosome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#47 |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) |
+Supramolecule #2: ribosome
| Supramolecule | Name: ribosome / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#47 |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) |
+Macromolecule #1: RNA (3341-MER)
| Macromolecule | Name: RNA (3341-MER) / type: rna / ID: 1 / Details: 25S rRNA / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 1.0799771249999999 MDa |
| Sequence | String: GGUUGACCUC GGAUCAGGUA GGAGGACCCG CUGAACUUAA GCAUAUCAAU AAGCGGAGGA AAAGAAACCA ACAGGGAUUG CCCUAGUAA CGGCGAGUGA AGCGGCAACA GCUCAAAUUU GAAAGCUGGC UUCGGCCCGC GUUGUAAUUU GGAGAGGAUG C UUUGGGCG ...String: GGUUGACCUC GGAUCAGGUA GGAGGACCCG CUGAACUUAA GCAUAUCAAU AAGCGGAGGA AAAGAAACCA ACAGGGAUUG CCCUAGUAA CGGCGAGUGA AGCGGCAACA GCUCAAAUUU GAAAGCUGGC UUCGGCCCGC GUUGUAAUUU GGAGAGGAUG C UUUGGGCG AGGCUCCUUC UGAGUUCCCU GGAACGGGAC GCCACAGAGG GUGAGAGCCC CGUAUAGUUG GAAGCCAAGC CU GUGUAAA GCUCCUUCGA CGAGUCGAGU AGUUUGGGAA UGCUGCUCAA AAUGGGAGGU AAAUUUCUUC UAAAGCUAAA UAC CGGCCA GAGACCGAUA GCGCACAAGU AGAGUGAUCG AAAGAUGAAA AGCACUUUGA AAAGAGGGUU AAAUAGCACG UGAA AUUGU UGAAAGGGAA GCGCUUGUGA CCAGACUUGC GCCCGGCGGA UCAUCCGGUG UUCUCACCGG UGCACUCCGC CGGGC UCAG GCCAGCAUCG GUUCUGGCGG GGGGAUAAAG GCCCAGGGAA UGUGGCUCCU CCGGGAGUGU UAUAGCCCUG GGUGUA AUA CCCUCGCCGG GACCGAGGAC CGCGCUCUGC AAGGAUGCUG GCGUAAUGGU CACCAGCGAC CCGUCUUGAA ACACGGA CC AAGGAGUCAA GGUUUUGCGC GAGUGUUUGG GUGUAAAACC CGCACGCGUA AUGAAAGUGA ACGUAGGUGA GAGCUUCG G CGCAUCAUCG ACCGAUCCUG AUGUAUUCGG AUGGAUUUGA GUAGGAGCGU UAAGCCUUGG ACCCGAAAGA UGGUGAACU AUGCUUGGAU AGGGUGAAGC CAGAGGAAAC UCUGGUGGAG GCUCGCAGCG GUUCUGACGU GCAAAUCGAU CGUCAAAUCU GAGCAUGGG GGCGAAAGAC UAAUCGAACC AUCUAGUAGC UGGUUACCGC CGAAGUUUCC CUCAGGAUAG CAGUGUCGAC C UUCAGUUU UAUGAGGUAA AGCGAAUGAU UAGGGACUCG GGGGCGAUUU UUAGCCUUCA UCCAUUCUCA AACUUUAAAU AU GUAAGAA GCCCUUGUUA CUUAACUGAA CGUGGGCAUU CGAAUGUAUC GACACUAGUG GGCCAUUUUU GGUAAGCAGA ACU GGCGAU GCGGGAUGAA CCGAACGCGG GGUUAAGGUG CCGGAGUGGA CGCUCAUCAG ACACCACAAA AGGCGUUAGU ACAU CUUGA CAGCAGGACG GUGGCCAUGG AAGUCGGAAU CCGCUAAGGA CUGUGUAACA ACUCACCUGC CGAAUGUACU AGCCC UGAA AAUGGAUGGC GCUCAAGCGU CCCACCCAUA CCCCGCCCUC AGGGUAGAAA CGAUGCCCUG AGGAGUAGGC GGCCGU GGA GGUCAGUGAC GAAGCCUAGG GCGUGAGCCC GGGUCGAACG GCCUCUAGUG CAGAUCUUGG UGGUAGUAGC AAAUACU UC AAUGAGAACU UGAAGGACCG AAGUGGGGAA AGGUUCCAUG UGAACAGCGG UUGGACAUGG GUUAGUCGAU CCUAAGCC A UAGGGAAGUU CCGUUUCAAA GGGGCACUCG UGCCCCGUGU GGCGAAAGGG AAGCCGGUUA AUAUUCCGGC ACCUGGAUG UGGGUUUUGC GCGGCAACGC AACUGAACGC GGAGACGACG GCGGGGGCCC CGGGCAGAGU UCUCUUUUCU UCUUAACGGU CUAUCACCC UGGAAACAGU UUGUCUGGAG AUAGGGUUUA AUGGCCGGAA GAGCCCGACA CUUCUGUCGG GUCCGGUGCG C UCUCGACG UCCCUUGAAA AUCCGCGGGA GGGAAUAAUU CUCACGCCAG GUCGUACUCA UAACCGCAGC AGGUCCCCAA GG UGAACAG CCUCUGGUUG AUAGAACAAU GUAGAUAAGG GAAGUCGGCA AAAUAGAUCC GUAACUUCGG GAAAAGGAUU GGC UCUAAG GGUUGGGCAC GUUGGGCUUU GGGCGGACGC CCUGGGAGCA GAGGGCCUCU AGCCGGGCAA CCGGCCGGCG GCCC UCAGC ACCCGGGGUU GAAGCCCUUA GCAGGCUUCG GCCGUCCGGC GUGCGGUUAA CAACCAACUU AGAACUGGUA CGGAC AGGG GGAAUCUGAC UGUCUAAUUA AAACAUAGCA UUGCGAUGGC CAGAAAGUGG UGUUGACGCA AUGUGAUUUC UGCCCA GUG CUCUGAAUGU CAAAGUGAAG AAAUUCAACC AAGCGCGGGU AAACGGCGGG AGUAACUAUG ACUCUCUUAA GGUAGCC AA AUGCCUCGUC AUCUAAUUAG UGACGCGCAU GAAUGGAUUA ACGAGAUUCC CACUGUCCCU AUCUACUAUC UAGCGAAA C CACAGCCAAG GGAACGGGCU UGGCAAAAUC AGCGGGGAAA GAAGACCCUG UUGAGCUUGA CUCUAGUUUG ACAUUGUGA AAAGACAUAG GAGGUGUAGA AUAGGUGGGA GCUUCGGCGC CAGUGAAAUA CCACUACUCC UAUUGUUUUU UUACUUAUUC AAUGAAGCG GGGCUGGACU UGCGUCCAAC UUCUGGAGUU AAGGUCCUUC GCGGGCCGAC CCGGGUUGAA GACAUUGUCA G GUGGGGAG UUUGGCUGGG GCGGCACAUC UGUUAAACCA UAACGCAGGU GUCCUAAGGG GGGCUCAUGG AGAACAGAAA UC UCCAGUA GAACAAAAGG GUAAAAGUCC CCUUGAUUUU GAUUUUCAGU GUGAAUACAA ACCAUGAAAG UGUGGCCUAU CGA UCCUUU AGUCCCUCGA AAUUUGAGGC UAGAGGUGCC AGAAAAGUUA CCACAGGGAU AACUGGCUUG UGGCGGCCAA GCGU UCAUA GCGACGUCGC UUUUUGAUCC UUCGAUGUCG GCUCUUCCUA UCAUACCGAA GCAGAAUUCG GUAAGCGUUG GAUUG UUCA CCCACUAAUA GGGAACGUGA GCUGGGUUUA GACCGUCGUG AGACAGGUUA GUUUUACCCU ACUGAUGAAC UCGUCG CAA UGGUAAUUCA GCUUAGUACG AGAGGAACCG CUGAUUCAGA UAAUUGGUUU UUGCGGUUGU CCGACCGGGC AGUGCCG CG AAGCUACCAU CUGCUGGAUA AUGGCUGAAC GCCUCUAAGU CAGAAUCCAU GCCAGAACGC GACGAUACUA CCCGCACG U UGUAGACGUA UAAGAAUAGG CUCCGGCCUC GUAUCCUAGC AGGCGAUUCC UCCGCCGGCC UCGAAGUGGC CGUCGGUAA UUCGCGUAUU GCAAUUUAGA CACGCGCGGG AUCAAAUCCU UUGCAGACGA CUUAGAUGUG CGAAAGGGUC CUGUAAGCAG UAGAGUAGC CUUGUUGUUA CGAUCUGCUG AGGGUAAGCC CUCCUUCGCC UAGAUUUCCC AG |
+Macromolecule #2: RNA (319-MER)
| Macromolecule | Name: RNA (319-MER) / type: rna / ID: 2 / Details: 5.8S rRNA / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 102.60743 KDa |
| Sequence | String: AAACUUUCAA CAACGGAUCU CUUGGUUCUG GCAUCGAUGA AGAACGCAGC GAAAUGCGAU AAGUAAUGUG AAUUGCAGAA UUCCGUGAA UCAUCGAAUC UUUGAACGCA CAUUGCGCCC GCCGGUAUUC CGGCGGGCAU GCCUGUUCGA GCGUCAUUUC A ACCAUCAA ...String: AAACUUUCAA CAACGGAUCU CUUGGUUCUG GCAUCGAUGA AGAACGCAGC GAAAUGCGAU AAGUAAUGUG AAUUGCAGAA UUCCGUGAA UCAUCGAAUC UUUGAACGCA CAUUGCGCCC GCCGGUAUUC CGGCGGGCAU GCCUGUUCGA GCGUCAUUUC A ACCAUCAA GCCCUGGGCU UGUGUUGGGG ACCCGCGGCU GCUCGCGGGC CCUGAAAAGC AGUGGCGGGC UCGCUAGUCA CA CCGAGCG UAGUAGAACU UCUAUUCGAU CUCGCUCAGG GCGUGCGGCG GGUGCCAGCC GUAAAACCCC AGCUUCUUCU AA |
+Macromolecule #3: Brix domain-containing protein
| Macromolecule | Name: Brix domain-containing protein / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 36.48202 KDa |
| Sequence | String: MAAVYKTIAK VTGSKEDKDK EDRSNIIAER KNKQRVLVLS SRGVTYRHRH LLNDLASMLP HGRKDAKFDT KSRLYELCEL AELYNCNNV LFFEARKGKD LYMWFSKVPN GPTVKFYAQN LHTMEELHFQ GNCLKGSRPI LSFDAAFEQE PYLKVIKELF L HTFGVPQG ...String: MAAVYKTIAK VTGSKEDKDK EDRSNIIAER KNKQRVLVLS SRGVTYRHRH LLNDLASMLP HGRKDAKFDT KSRLYELCEL AELYNCNNV LFFEARKGKD LYMWFSKVPN GPTVKFYAQN LHTMEELHFQ GNCLKGSRPI LSFDAAFEQE PYLKVIKELF L HTFGVPQG HKKSKPFIDH VLSFSVADGK IWVRNYEIRE VEKVKGEADK REEGETEKPK AAKPGSKDTD INLIEIGPRF VL TPIIIQE GSFGGPILYE NKRFISPNKI RAELRKAKAA RHHARMEQQR DLLARKRQLG LDESTPKKKD DLDTRELFA UniProtKB: Brix domain-containing protein |
+Macromolecule #4: Ribosome biogenesis protein C8F11.04
| Macromolecule | Name: Ribosome biogenesis protein C8F11.04 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 43.700527 KDa |
| Sequence | String: MAPSTAVAKK ETDAVIPVDP DQTLKACKAL LAHIKKAAAA PRPDGKQNLL ADEESTVAET PIWLTLTTKK HIHDSHRLQP GKIILPHPL NTSEEISVCL ITADPQRFYK NAVADEFPED LRAKIGRVID ISHLKAKFKA YEAQRKLFSE HDVFLADTRI I NRLPKALG ...String: MAPSTAVAKK ETDAVIPVDP DQTLKACKAL LAHIKKAAAA PRPDGKQNLL ADEESTVAET PIWLTLTTKK HIHDSHRLQP GKIILPHPL NTSEEISVCL ITADPQRFYK NAVADEFPED LRAKIGRVID ISHLKAKFKA YEAQRKLFSE HDVFLADTRI I NRLPKALG KTFYKTTTKR PIPVVLMAQR EKVNGKRVPA PKGKKEKRDP LENANARPIP EIVAEIRKAI GAALVHLSPS TN TAIKVGY ANWEPEKLAA NIETVIRELV ERFVPQKWQN VRNFYVKGPE TAALPIYQTD ELWLDESKVV PDGQEPARAL PGK REKANI GKKRKPLEDA SQPALEETGK DERPKKKAKK TLPESNDDKL DKAIAERKEQ LKKQKAAAKK VAADI UniProtKB: Uncharacterized protein |
+Macromolecule #5: Ribosome biogenesis protein ERB1
| Macromolecule | Name: Ribosome biogenesis protein ERB1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 90.838023 KDa |
| Sequence | String: MGSKIVEKKR KSRDSDSESD NELGDGLFDG VLSQSEDEED YIPSSEVDED DDDDADESAS EDSDDSNDSE DDEVEEDDEA LLSDEIPSE GESEKDQDLA ESKESKQDQD KEPSEPEILE PFVDPPRKED EELEDRNYRI EKDANGGIRY VYDEIDPVYD S DDTDYNVP ...String: MGSKIVEKKR KSRDSDSESD NELGDGLFDG VLSQSEDEED YIPSSEVDED DDDDADESAS EDSDDSNDSE DDEVEEDDEA LLSDEIPSE GESEKDQDLA ESKESKQDQD KEPSEPEILE PFVDPPRKED EELEDRNYRI EKDANGGIRY VYDEIDPVYD S DDTDYNVP VNTIGNIPLS FYDSYPHIGY DINGKKIMRP ATGDALQNLL DSIEVPEGWT GLTDPNTGKP LNLSRDELEL IR KVQQGLI PDDVEDPYPD TVEWFTSVEE KMPLSAAPEP KRRFIPSKNE AKQIMKLVRA IREGRILPYK PPEEREREEL EKE EEFYDL WQNEEPQPPN PMHIPAPKLP PPGYDLSYNP PPEYLPTKEE REEWEKMDPE DREKDYLPTK YDSLRKVPAW GNFV KERFE RCMDLYLAPR VRKNRLNIDP NSLLPKLPSP DELKPFPTVQ QTIFRGHEGR VRSVAIDPTG VALATGGDDG TVRVW ELLT GRQVWSVKLN GDEAVNTVRW RPTKDTFILA AAAGEDIFLM IPTHPSVTPA LDQASRDILN AGFGHATNGK QQANLP PGK EPPGKWARPG TRLEDEGVLL RITVRSTIKA ISWHRRGDHF ATVSPSGQRS SVAIHTLSKH LTQIPFRKLN GLAQTAS FH PLRPLFFVAT QRSIRCYDLQ KLELVKIVQP GAKWISSFDV HPGGDNLVVG SYDKRLLWHD LDLSNRPYKT MRFHTEAI R AVRFHKGGLP LFADASDDGS LQIFHGKVPN DQLENPTIVP VKMLKGHKVV NKLGVLDIDW HPREPWCVSA GADGTARLW M UniProtKB: Ribosome biogenesis protein ERB1 |
+Macromolecule #6: Ribosome biogenesis protein YTM1
| Macromolecule | Name: Ribosome biogenesis protein YTM1 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 52.989242 KDa |
| Sequence | String: MDAPMEDAPA PVAQVKVIFT TTEPDLELPE SKRQLLVPAD IRRYGLSRIL NSESMLDTGS IPFDFLINGS FLRSSLEDYL TSNGLSLDT TLTLQYVRSL IPPVYEASFE HDDWVSAVDV LSATSPAGRW SSAANSSAAV QPGQERVLSA SYDGLLRIWN A SGSVIATS ...String: MDAPMEDAPA PVAQVKVIFT TTEPDLELPE SKRQLLVPAD IRRYGLSRIL NSESMLDTGS IPFDFLINGS FLRSSLEDYL TSNGLSLDT TLTLQYVRSL IPPVYEASFE HDDWVSAVDV LSATSPAGRW SSAANSSAAV QPGQERVLSA SYDGLLRIWN A SGSVIATS PSGSHGGHTA SIKAAKFLTS DRLASAGMDR TVRVWKYTES DHFTGELKPT LELYGHTGSV DWLDVDGHSK HI LTASADG AIGFWSASKA SAPEPDASLL PGAHVSKRRK ATSSVSTAQR GPLGLWSIHT APATAAIFDP RDRTVAYSAS QDH TVRTLD LTTGQVVSTL TLTHPLLSLS ALTRAGTTSP LLAAGTSARH ITMVDPRASS ATTSVMTLRG HANKVVSLSP SPEN EYSLV SGSHDGTCRV WDLRSVRPAT KEEGSLGGVS EPVYVIERES WASKGKKKRP VAGDGCKVFS VVWDKLGIFS GGEDK KVQV NRGRNIVTEQ K UniProtKB: Ribosome biogenesis protein YTM1 |
+Macromolecule #7: RNA helicase
| Macromolecule | Name: RNA helicase / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 66.830867 KDa |
| Sequence | String: MASESSKKRK FKDSKGPKPD EAVAKPSKFK KLKRDPTPEE RETSDSESGN SDGQHEPDQV DSDGEDAVSD DGSFDEKVAH QDEDGSRPE NGTDLEDVPK NDNLLPPPTN AQEFSELNLS EKTTKAIAEM GFTKMTEIQR RAIPPALAGK DVLGAAKTGS G KTLAFLIP ...String: MASESSKKRK FKDSKGPKPD EAVAKPSKFK KLKRDPTPEE RETSDSESGN SDGQHEPDQV DSDGEDAVSD DGSFDEKVAH QDEDGSRPE NGTDLEDVPK NDNLLPPPTN AQEFSELNLS EKTTKAIAEM GFTKMTEIQR RAIPPALAGK DVLGAAKTGS G KTLAFLIP AVEMLSSLRF KPRNGTGAIV VTPTRELALQ IFGVARELMK YHSQTYGVVI GGANRRAEAE KLGKGVNLLI AT PGRLLDH LQNTPFVFKN LKSLIIDEAD RILEIGFEDE MRQIVKILPK EDRQTMLFSA TQTTKVEDLA RISLRPGPLY INV DEEKKY STVEGLEQGY VVVEADKRFL LLFSFLKKMA KKKIIVFFSS CNSVKYYSEL LQYIDLPVLD LHGKQKQQKR TNTF FEFCN AKSGTLICTD VAARGLDIPQ VDWIVQFDPP DDPRDYIHRV GRTARGNNGK GRSLLFLQPC ELGFLAHLKA AKVPV VEYD FPKNKILNVQ SQLEKLISTN YYLNQSAKEG YRSYIHAYAS HSLRSVFDVH KLDLVKVAKS FGFSTPPRVD ITLSAS LSR DKKPQGRRAY GSQPRQGGRY KKVGDIPDPQ PNTFG UniProtKB: ATP-dependent RNA helicase |
+Macromolecule #8: Ribosome assembly factor mrt4
| Macromolecule | Name: Ribosome assembly factor mrt4 / type: protein_or_peptide / ID: 8 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 30.551549 KDa |
| Sequence | String: MPKSKRARVY HLIQVNKKGR EAKERLFSNI RETIPKYQHC FVFSVDNMRN NYLKDVRHEL NDCRIFFGKT KLMARALGTT PEEEQADGL HRLTRYLTGT VGLLFTNRDP ADIESYFSNL SQVDFARAGT VAPRTVTVPT GIVYSTGGEV PPEHDVPVSH T LEPELRRL ...String: MPKSKRARVY HLIQVNKKGR EAKERLFSNI RETIPKYQHC FVFSVDNMRN NYLKDVRHEL NDCRIFFGKT KLMARALGTT PEEEQADGL HRLTRYLTGT VGLLFTNRDP ADIESYFSNL SQVDFARAGT VAPRTVTVPT GIVYSTGGEV PPEHDVPVSH T LEPELRRL GMPVRMIKGK VCLGDEKGEA SEGYTICKEG EVLDSRQTRL LKLFSICLSE FKVSLLGYWN SASGEVTELE AG KTRPKRE GNRRQAMNGD EIDEDQSSDE DSD UniProtKB: Large ribosomal subunit protein uL10 |
+Macromolecule #9: 60S ribosome subunit biogenesis protein NIP7
| Macromolecule | Name: 60S ribosome subunit biogenesis protein NIP7 / type: protein_or_peptide / ID: 9 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 20.685982 KDa |
| Sequence | String: MRPLTDQEMK IVLDKLANYM TDLKSLIAPL EDGDRYVFRM QKDRVYYVKL SIANIATCVA RDKLLSLGTC LGKMTKSGKF RLHITALPI LAQNARYKIW VKDNGAQPFL YGSNIVKAHV GRWTEDCPEH SGCVVYNMAD IPLGFGVTAR STAEARRLDP T GIVCFRQA DCGEYLRDED TLFTGA UniProtKB: 60S ribosome subunit biogenesis protein NIP7 |
+Macromolecule #10: Nucleolar GTP-binding protein 1
| Macromolecule | Name: Nucleolar GTP-binding protein 1 / type: protein_or_peptide / ID: 10 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 75.621508 KDa |
| Sequence | String: MTGWKDIPPV PTAQEFIDIV LSRTQRRLPT QIRPGFKISR IRAFYTRKVK FTQETCSEKF GAIISSFPVL SDQHPFHRDL MNILYDADH FKVALGQIST AKNLIETISR DYVRLLKYAQ SLYQCKQLKR AALGRMATLI KRLKDPLIYL DQVRQHLARL P DINPTTRT ...String: MTGWKDIPPV PTAQEFIDIV LSRTQRRLPT QIRPGFKISR IRAFYTRKVK FTQETCSEKF GAIISSFPVL SDQHPFHRDL MNILYDADH FKVALGQIST AKNLIETISR DYVRLLKYAQ SLYQCKQLKR AALGRMATLI KRLKDPLIYL DQVRQHLARL P DINPTTRT LLVAGFPNVG KSSFVRSVTR ADTPVEPYAF TTKSLFVGHL DYKYLRYQVI DTPGILDHPL EEMNTIEMQS VT ALAHLRA AVLYFMDISE QCGFSLKAQI NLFKSIKPLF ANKMVFIVLN KMDIKKFEEL DPEMQQEIND LTKSGEVEIL RAS CATQEG VQEVKNHVCE RLLVERVSQK LKAGTHSNGN IGTRLQEVMA RIHVATPMDG TTRETFIPEA VKNLKKYDKN DPNR RVLAR DIEEANGGAG VFNVDLRKDW ILENPEWKYD KIPEIFDGKN VYDYIDPDID AKLQALEEEE ERLEKEGFYD EDDDL PDEE EEEILQKAEY IREQHALIRN EAKMRKSLKN RAIIPRKAVK KPLSQLEDHL DQLGVDTEAI GLRARAQTAQ SSRGRS LVR SRGTTADPDA MDIDDGAASA KERLARSRSR ARSVAATNRL QDGVQGTTLR SKAERQAKLA QRKMNRMARQ GEADRHI HA SMPKHLFSGK RTIGKTDRR UniProtKB: Nucleolar GTP-binding protein 1 |
+Macromolecule #11: Putative RNA-binding protein
| Macromolecule | Name: Putative RNA-binding protein / type: protein_or_peptide / ID: 11 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 45.884414 KDa |
| Sequence | String: MAAELRKKKS KSAVADADAA ISKKEKATKV TKATPKAEKR KAPEEASPVA IKKQKADKDA VVKKAAVAAE DKIAEKPVKS VLKKETSAK AKEAAKPKKK QEKVEEEEEE ALENADAAFG SDEESELDEQ TKALMDTLDD GEESDEDESK QVSTFKKGQD V GKIPTPKK ...String: MAAELRKKKS KSAVADADAA ISKKEKATKV TKATPKAEKR KAPEEASPVA IKKQKADKDA VVKKAAVAAE DKIAEKPVKS VLKKETSAK AKEAAKPKKK QEKVEEEEEE ALENADAAFG SDEESELDEQ TKALMDTLDD GEESDEDESK QVSTFKKGQD V GKIPTPKK APQKQTNGDS KPAGDSQPGV MYIARLPHGF YEHELRGYFS QFGEITRLRV VRNKKTGASR HRAFIEFADA EV ADIAART MDKYLLFGHI LTCKIVPPAQ VHPDLFKGAN RRFKVVPWNK MAGRQLERPL SESQWQVKVA KEEQRRAARA EKL KEMGYE FEAPALKVPK AKPVLENGEE EKKAIETAPA PAVEEKESKE VAKRQAEEEV EEKAPAPKTK KVKEDKPAKE AATI SAPKK GKKSKKTKS UniProtKB: Putative RNA-binding protein |
+Macromolecule #12: Pescadillo homolog
| Macromolecule | Name: Pescadillo homolog / type: protein_or_peptide / ID: 12 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 77.16725 KDa |
| Sequence | String: MGKAKKKGKS GAARNYMTRT QAVKKLQLSL PDFRKLCIWK GIYPREPRDR RKVNKSATAS TTFYYTKDIQ YLLHEPLLQK FREQKALEK KISRALGRGD VSNAARLERN ANLPEKTGKP RYTLNHIIRE RYPTFQDALR DLDDCLSMLF LFANLPSTTA V PAKMIARC ...String: MGKAKKKGKS GAARNYMTRT QAVKKLQLSL PDFRKLCIWK GIYPREPRDR RKVNKSATAS TTFYYTKDIQ YLLHEPLLQK FREQKALEK KISRALGRGD VSNAARLERN ANLPEKTGKP RYTLNHIIRE RYPTFQDALR DLDDCLSMLF LFANLPSTTA V PAKMIARC ERLCHEFQHY LIVTHSLRKS FLSIKGIYYQ ANIQGEDILW LVPYKFNQRI VGDVDFRIMG TFVEFYMTLL GF VNYRLYT SIGLKYPPKF DQVKDDQGAE LAAFSLEGLN LASQNGEQKA ITNGEEHGPD PKVQAEVDKL VAKLREEEQQ ANG DKTDEK GEENEGDDKP SDAIDKFEPV APGGDVLPQP SYSSSDPSQL FANFTFFLSR ETPRQPLEFI LRAFGCKRIG WDAV LGEGA FTTDESDPRI THQIIDRPVI RAAVSEDGDG EDNQTSQKLA PNGRYPGRIY VQPQWVWDSI NDEELKPPEL YAPGA QLPP HLSPFVKPTQ GQYDPTKPLE EQQTEAEALE AELEDAQAQQ EGSDEESGSE VENDMSVASD EEDEQEDDFG GFSDED EEQ SDEGSEEGDE EEDDDEEEEA TLERQRELEA ELAGKAVSKG KPLDPKVKAK LEAKKALERK KKQEAEELER AKGMLSK KK RKLFEQMQYS NAKKNAEDAK LRAKRRRIEK EMAAKKA UniProtKB: Pescadillo homolog |
+Macromolecule #13: Ribosome biogenesis protein NSA2 homolog
| Macromolecule | Name: Ribosome biogenesis protein NSA2 homolog / type: protein_or_peptide / ID: 13 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 29.72458 KDa |
| Sequence | String: MPQNEYIERH RKLHGRRLDA EERARKKAAR EGHKNSENAQ NLRGLRAKLY AKQRHAQKIQ MRKAIKQHEE RNVKGRPAEK EPSDPIPSY LLDRANPTTA KALSSQIKNK RAEKAARFSV PIPKVRGISE EEMFKVVKTG KKTHKKGWKR IVTKPTFVGP D FTRRPVKY ...String: MPQNEYIERH RKLHGRRLDA EERARKKAAR EGHKNSENAQ NLRGLRAKLY AKQRHAQKIQ MRKAIKQHEE RNVKGRPAEK EPSDPIPSY LLDRANPTTA KALSSQIKNK RAEKAARFSV PIPKVRGISE EEMFKVVKTG KKTHKKGWKR IVTKPTFVGP D FTRRPVKY ERFIRPMGLR YKKANVTHPT LNVTVQLPIL SVKKNPSNPL YTQLGVLTKG TIIEVNVSDL GIVTASGKIA WG RYAQITN NPENDGCVNA VLLV UniProtKB: Ribosome biogenesis protein NSA2 homolog |
+Macromolecule #14: Putative GTP binding protein
| Macromolecule | Name: Putative GTP binding protein / type: protein_or_peptide / ID: 14 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 61.523926 KDa |
| Sequence | String: MAGTINKPKK PTSKRKTTRL RAKISKRAAE KKRKERKLAR KNPEWRSKLK KDPGIPNLFP YKERLLQQRE EERIRRKEEL LRKKELAKA AKTGADGDNK DDEQMQEDDE HMIREEELDL DDAMDEDGSD VDESNPLAAL VRSARKAAEE YEKELQSGSE M DEDDEDDS ...String: MAGTINKPKK PTSKRKTTRL RAKISKRAAE KKRKERKLAR KNPEWRSKLK KDPGIPNLFP YKERLLQQRE EERIRRKEEL LRKKELAKA AKTGADGDNK DDEQMQEDDE HMIREEELDL DDAMDEDGSD VDESNPLAAL VRSARKAAEE YEKELQSGSE M DEDDEDDS DDASDAGSDA GVITEVHGGA TSRKAYDKVF KQVVEQADVI LYVLDARDPE GTRSHDVEQA VMAAAGGGKR LM LILNKVD LVPPPVLKGW LTYLRRFFPT LPLRASNPAP NARTFSHRDI TVQSTSAALF RALKAYAAAR NLKRAIAVGV IGY PNVGKS SVINALLSRL PGSARGGRTP CPAGAEAGVT TAIRAVKIDS KLTLLDSPGI VFPSTASSQT FIPKNPVEAH AHLV LLNAI PPKQIEDPVP AVTLLLKRLS ATPELMDRLM QVYDIPPLLK DPSQGGDATM DFLVQVARKR GRLGRGGVPN IQAAA MTVV TDWRDGRIQG WTEPPKIAVE GVKEGNEGKV VRKIADQEVA PDQKIIVTEW AKEFKLAGLW GDEEQTEEGD KMEA UniProtKB: Putative GTP binding protein |
+Macromolecule #15: 60S ribosomal protein l7-like protein
| Macromolecule | Name: 60S ribosomal protein l7-like protein / type: protein_or_peptide / ID: 15 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 28.821816 KDa |
| Sequence | String: MSSTTVPTQN DILVPETLLK KRKSQEKARA ERAAALEKRK QANKEKRQVI FKRAEKYVKE YREQEREKIR LARIAKQQGS FHIPAEAKL VFVIRIKGIN KIPPKPRKIL QLLRLRQINN GVFVKVTKAT AEMIKIVEPW VAYGYPNLKS VRELIYKRGY G KVNGQRIP ...String: MSSTTVPTQN DILVPETLLK KRKSQEKARA ERAAALEKRK QANKEKRQVI FKRAEKYVKE YREQEREKIR LARIAKQQGS FHIPAEAKL VFVIRIKGIN KIPPKPRKIL QLLRLRQINN GVFVKVTKAT AEMIKIVEPW VAYGYPNLKS VRELIYKRGY G KVNGQRIP LTDNAIIEEN LGKYGIICIE DLIHEIFTVG PNFKQAANFL WPFKLSNPNG GFRPRKFKHF IEGGDLGNRE EH INALIRA MN UniProtKB: 60S ribosomal protein l7-like protein |
+Macromolecule #16: Eukaryotic translation initiation factor 6
| Macromolecule | Name: Eukaryotic translation initiation factor 6 / type: protein_or_peptide / ID: 16 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 26.46884 KDa |
| Sequence | String: MAVRAQFENS NEVGVFATLT NSYCLVALGA SENFYSVFEA ELQDVIPICR TTIAGTRIIG RLTAGNRKGL LVPTTTTDQE LQHLRNSLP DDIRIQRIEE RLSALGNVIV CNDHTALIHP DLERETEEII ADVLGVEVFR QTIADHVLVG SYMALSNQGG L VHPKTSIQ ...String: MAVRAQFENS NEVGVFATLT NSYCLVALGA SENFYSVFEA ELQDVIPICR TTIAGTRIIG RLTAGNRKGL LVPTTTTDQE LQHLRNSLP DDIRIQRIEE RLSALGNVIV CNDHTALIHP DLERETEEII ADVLGVEVFR QTIADHVLVG SYMALSNQGG L VHPKTSIQ DQDELSSLLG VPLVAGSVNR GSNVIGGGMV VNDWLAVTGL DTTAPELSVI ESVFRLGEGA GPGAINTSMK NT IVESFY UniProtKB: Eukaryotic translation initiation factor 6 |
+Macromolecule #17: DUF2423 domain-containing protein
| Macromolecule | Name: DUF2423 domain-containing protein / type: protein_or_peptide / ID: 17 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 13.262389 KDa |
| Sequence | String: MAKSARASRI KENHQRFKKN IAGPVEAARL ERLSAKLMAI AQAPKPDIEM KEESEATAEQ PKESTQTKDD TAMDVDGAKP TLSGVKSGK SIGRKDSRKR RGKKSSIVFP MKGPKRNNLK K UniProtKB: DUF2423 domain-containing protein |
+Macromolecule #18: RNA methyltransferase nop2-like protein
| Macromolecule | Name: RNA methyltransferase nop2-like protein / type: protein_or_peptide / ID: 18 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 83.658305 KDa |
| Sequence | String: MGTKRRLKHQ GPPEPLPEEH FLKLKRKKGL PVEDTTPAEA PSAKKRRTST KEKETKPHKA TTKQTNGTKK APGAPSNGEK ASKPKPKKL PESEPESDVE MDDEFGGSDL DDVSISGSDT SEPKLGSDFM ASDDDSVYDS EQDEGPGKKT KFVFSDDEDE D EDEIEQKL ...String: MGTKRRLKHQ GPPEPLPEEH FLKLKRKKGL PVEDTTPAEA PSAKKRRTST KEKETKPHKA TTKQTNGTKK APGAPSNGEK ASKPKPKKL PESEPESDVE MDDEFGGSDL DDVSISGSDT SEPKLGSDFM ASDDDSVYDS EQDEGPGKKT KFVFSDDEDE D EDEIEQKL TAANIEGLSR KLDEQLAREA EENEEEIRNQ ALQTNIDGDK PKILEGNEED ELAAKTNGLL APDLQMLRTR IT DTIRVLE DFQNLAEEGR SRAEYTNQLL KDICAYYGYN EYLAEKLLNL FPPREAFAFF EANETPRPVV IRTNTLRTHR RDL AQALIN RGVTLEPVGK WSKVGLQVFD SKVPLGATPE YLAGHYILQA ASSFLPVMAL CPQENERCLD MAAAPGGKTT HMAA LMKNT GVIFANDPSK SRAKGLIGNI HRLGVRNTIV CNYDAREFPR VIGGFDRVLL DAPCSGTGVI CKDPSVKTNR DAKDF MQLP HTQKQLLLAA IDSCNHASKT GGYIVYSTCS VCVEENEEVV NYALSRRPNV KLVETGLPFG KEGFTSYMGK TFHPSL KLT RRFYPHLYNV DGFFVAKFKK IGPTPPNAVL ANGKKDQPKP ANATGEEEII DKTPIGAEEQ KKEDDFGGWD DEEDKVY ME RAKRNAMRRR GLDPRALKKP QGKKDDKDGE ASKEGANGET KKAATEQKKT EEKKTEEKKA EEKKAEKAKE EKPKAKAE K TTEKAKEGAK EKVKEQNKAN DKKEKAKGKK UniProtKB: Nucleolar protein 2 |
+Macromolecule #19: Ribosome biogenesis protein RLP24
| Macromolecule | Name: Ribosome biogenesis protein RLP24 / type: protein_or_peptide / ID: 19 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 26.434312 KDa |
| Sequence | String: MRIENCFFCG RPAWPSKGIT FMRNDGKAFR FCRSKCHKNF KMKRNPRKLK WTKAYRKNAG KEMVVDSTLQ FAARRNVPVR YNRELWQKT LKAMERISEI RARRERVFYK KRMAGKRARE VAAARKLVAE NEHLLPRLRG SEKRRLAELA AERGVDVEEL E REELAKLA ...String: MRIENCFFCG RPAWPSKGIT FMRNDGKAFR FCRSKCHKNF KMKRNPRKLK WTKAYRKNAG KEMVVDSTLQ FAARRNVPVR YNRELWQKT LKAMERISEI RARRERVFYK KRMAGKRARE VAAARKLVAE NEHLLPRLRG SEKRRLAELA AERGVDVEEL E REELAKLA SKKKSKAFGG EVRRVRVRTD GGVEEITESF GGKNVDEEMD DDDHDDSDRD DDDMDTD UniProtKB: Ribosome biogenesis protein RLP24 |
+Macromolecule #20: Nucleolar protein 16
| Macromolecule | Name: Nucleolar protein 16 / type: protein_or_peptide / ID: 20 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 26.960477 KDa |
| Sequence | String: MGRELQKRKR RSSRPTIRMP NRRKKALNPA GNNIIAKSWN KKETLSQNYT RFGLVAKLGK ATGGTAPGNK ALLKDLNDPL AIKSGDQGL IKVSEVKVER DEQGRIKRVV RDNNPLNDPL NDLDSDSESD AVPQQQQQEQ PVLNNHIRQH DLELENVAAD V DDESKPEV ...String: MGRELQKRKR RSSRPTIRMP NRRKKALNPA GNNIIAKSWN KKETLSQNYT RFGLVAKLGK ATGGTAPGNK ALLKDLNDPL AIKSGDQGL IKVSEVKVER DEQGRIKRVV RDNNPLNDPL NDLDSDSESD AVPQQQQQEQ PVLNNHIRQH DLELENVAAD V DDESKPEV LRALEREATR PVEKTVRHQS EREREWLQRL VDKHGDDVAA MARDRKLNPY QQTASDIKRR LKKAGLLSA UniProtKB: Nucleolar protein 16 |
+Macromolecule #21: AdoMet-dependent rRNA methyltransferase SPB1
| Macromolecule | Name: AdoMet-dependent rRNA methyltransferase SPB1 / type: protein_or_peptide / ID: 21 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 95.510961 KDa |
| Sequence | String: MAKTQKKHGK GRLDKWYKLA KEKGYRARAA FKLIQLNKKY GFLEKAKVVL DLCAAPGSWC QVCAETMPKD SLIIGVDLAP IKPIPKVIT FQSDITTEKC RATIRSHLKT WKADVVLHDG APNVGTAWVQ DSYNQAELAL HSLKLATEFL IEGGTFVTKV F RSKDYNKL ...String: MAKTQKKHGK GRLDKWYKLA KEKGYRARAA FKLIQLNKKY GFLEKAKVVL DLCAAPGSWC QVCAETMPKD SLIIGVDLAP IKPIPKVIT FQSDITTEKC RATIRSHLKT WKADVVLHDG APNVGTAWVQ DSYNQAELAL HSLKLATEFL IEGGTFVTKV F RSKDYNKL LWVCNQLFTK VEATKPPSSR NVSAEIFVVC RGFKAPKRID PRLLDPRSIF EDLADPAPNN EARVYNPEQK KR KREGYEE GDYTQYKETS AIEFINTTDP IAILANYNKL SFEQPPNGDV ALAALEKLPE TTKEIRACCD DLKVLGKKDF RLL LKWRLR VREIFGLPSK KTAKQQPLVE EEAKVEPMDE ELKIQEELER IKEKERAKKK RERRKENERK HKEIVRMQMH MTTP MDIGM EQEGPRGDGA FFRLKEIDQT DALRRIAKGK MAMLTEAEQA DRDSGIGLSP DTDDDSDEDG DQLERELDAM YEHYK ERKA SQDAKYRAKR ARQEVDDEEW EGLSADEKGS DVDMDDDDSE RLEEDSSDEE AWSDDEELDT SKPLIKDLSG APQDGS GLS KRARGFFSQD VFQKIPGLLD TVSESEEEQD SDDNDGIPSI QEQKRMQKEK KEKEAKKQDD DDFEVVKEHD SDDDWEE KE KKSKDGRPNI DIITAEAMTL AHQLATGEKT KADLIDEGYN KYAFKQKEGL PDWFLEDEAK HDKPIKPITK EAAQAIKE K LRALNARPIK KVAEARARRK LRQAKKLEKL KKKADLLVDE EGMSEKEKAT TIAKMLSAAA RKKRRPQVKV VKATGANRG IKGRPKGVKG RYKMVDGRMK KEMRALKRLA KKKR UniProtKB: Uncharacterized protein |
+Macromolecule #22: Nucleolar complex-associated protein 3
| Macromolecule | Name: Nucleolar complex-associated protein 3 / type: protein_or_peptide / ID: 22 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 77.953883 KDa |
| Sequence | String: MATRPLKRRK VTPPRDDGQD AAEREAKIQK AFLKNAASWD LEEAWESRPR KGKKKDKKKD EERPGRLPIR TEQGTWQVQD DVESIPSDA EWLDSDGELA VEQQLEPVAP APAPEPPKIS EKEQIRQAQE ELAKIATQLN ENPEEYPGHF KALARIGETP I LAIQKLCI ...String: MATRPLKRRK VTPPRDDGQD AAEREAKIQK AFLKNAASWD LEEAWESRPR KGKKKDKKKD EERPGRLPIR TEQGTWQVQD DVESIPSDA EWLDSDGELA VEQQLEPVAP APAPEPPKIS EKEQIRQAQE ELAKIATQLN ENPEEYPGHF KALARIGETP I LAIQKLCI VTQMAVYKDV IPGYRIRPLG EDAVNEKVSK EVKRLRTYEQ ALVAGYHGYL KTLATYAASS IPEDRKGEPI SS IAFTCAC ELVNAVPHFN FRGDLLRILV KKLSTRKIDR DFVKCREALE KLFQDDEEGN ASQEAVSLLS KMMKAREYRV DES VLNLFL HLRLLSEFVG KGSRDTVIRP GDDRPIGKKP KKKWEFRTKK QRKLLKAEKE AQKVMEQADA TVSHEERERI QSEI LKMVF ATYFRILKAR VPHLMGAVLE GLAKYAHLIN QDFFGDLLEA LKDLIRDTDR LEEEDGPDSS QDDDQGVSVV RDTSR ESLL CTVTAFALLE GQDAHNARSD LHLDLSFFIT NLYRSLLSLS LNPDLELGAQ SLHLSDPNDT SSGSTENSRR NNKINL QTT TVLLLRCLTS VLLPPWNIRS VPPIRLAAFC KQLMTLALQV PEKSSQAILG LLQDVVHTHG RKVAALWNTE ERKGDGT YK PLSETVEGSN PFTTTIWEGE LLRKHYCPKV REGLKAMEKE LRSIPS UniProtKB: Nucleolar complex-associated protein 3 |
+Macromolecule #23: rRNA-processing protein EBP2
| Macromolecule | Name: rRNA-processing protein EBP2 / type: protein_or_peptide / ID: 23 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 50.255562 KDa |
| Sequence | String: MAKKGATQSP KAGSKPNSQT GDAQSKNQKA DRRKSNEPVV EKVAEKEEQR EEETEESEED ESEDEQLQIR ANGNNESDSE EDSDENSED EDESDDDSGD EHPTHHIDFE AIDDSDSESE LDSDEENDKK QDSNKAQNGM SEDKMDVDKD GEDKSGDEED E EEEIDVEE ...String: MAKKGATQSP KAGSKPNSQT GDAQSKNQKA DRRKSNEPVV EKVAEKEEQR EEETEESEED ESEDEQLQIR ANGNNESDSE EDSDENSED EDESDDDSGD EHPTHHIDFE AIDDSDSESE LDSDEENDKK QDSNKAQNGM SEDKMDVDKD GEDKSGDEED E EEEIDVEE LEPEDEEDEE QIAAGTRTRQ TINNKEGLLT ALRRIQLDIN PKTTPFSFHQ SVVASKRTED SIPQIEDDLQ RE LAFMNQA LEAARLGRSL LRKEGVPFTR PTDYFAEMVR TDATMEKVKQ KLIEEATAKK AAAEARKQRD LKKFGKAVQV QKQ LERAKQ KREALDKINQ LKRKRAEGGS AALGATEADD PFDVAVDNEL SRDSKPKKRR ADGDHPVPNP KRQKKNAKYG FGGK KRGSK SGDAISSGDL SGFSVKRMKY GGGAGGGKKK ANRPGKARRK AAAAKR UniProtKB: rRNA processing protein |
+Macromolecule #24: Putative 60S ribosomal protein
| Macromolecule | Name: Putative 60S ribosomal protein / type: protein_or_peptide / ID: 24 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 15.960451 KDa |
| Sequence | String: MSNVSADLIW EVSRNYNSFL VKQRTGTFSR DALNLTNQHS RKHAGFVNDK ALGIVPAEKG VKVIAKKVKA ANKPASSLYT VTYKSTARK AYKAIASQAA KHGYRADLRQ AAVARASAIL RSQRPVKPEP PKKLRGAAAR RAAAAGKQ UniProtKB: Putative 60S ribosomal protein |
+Macromolecule #25: ATP-dependent RNA helicase
| Macromolecule | Name: ATP-dependent RNA helicase / type: protein_or_peptide / ID: 25 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 75.468516 KDa |
| Sequence | String: MAPEQARSKK SSRSWTAFNP PLPQWILDYV SSMGYEQPTP VQKSCLDIFR GNKDVVVEAV TGSGKTLAFL IPVVERLLRL DEAAKKHHV QAIIISPTRE LASQIYNVLV SLIKFHPESS ELLQYAKSDE KRPATTKPVI VPQLLVGGTV KAAEDLSIFL R LSPNLLVG ...String: MAPEQARSKK SSRSWTAFNP PLPQWILDYV SSMGYEQPTP VQKSCLDIFR GNKDVVVEAV TGSGKTLAFL IPVVERLLRL DEAAKKHHV QAIIISPTRE LASQIYNVLV SLIKFHPESS ELLQYAKSDE KRPATTKPVI VPQLLVGGTV KAAEDLSIFL R LSPNLLVG TPGRLAELLS SPYVKTPASS FEVLVMDEAD RLLDLGFSPE LTRILGYLPK QRRTGLFSAS LSDAVERLIT VG MLYPHKI TVRVKSLKTG GVIEERKTPM SLQMSYIVTP ASHKIPALCQ ILEKLDPRPQ RSIVFFSTCF AVKYFARVLH GIL PAGYSI VSLHGKLEPH VREKNYERFV TATSPTVLLT TDIAARGLDI PQVDLVIQHD PPTDTKVFIH RCGRAGRAGR RGLS VVMLQ PGREEGYVQL LEVRQTPITP LEHPQISVTD TQADDVANKI REQAKKDREV FQLAQRAFVS WARSYMEHQA TSIFR VADL DWFDLAKGYG LLELPKMPET RGLDRSLGLG IDTSAIPFKD KAREKKRLAE LEKWKAEKAE KEAAAAAAGE TAGADG SNP LKRKKNEAWS GKHEQEELRQ LRKEKKRRKK EALKMARMTE KEKEELRKLE ELINEVRKRN QQAAAGTANG QPMKEGG SS SSSSSSSSSS NGTESKGGGK ESKGGADEES EFEGFDD UniProtKB: ATP-dependent RNA helicase |
+Macromolecule #26: 60S ribosomal subunit-like protein
| Macromolecule | Name: 60S ribosomal subunit-like protein / type: protein_or_peptide / ID: 26 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 22.678574 KDa |
| Sequence | String: MGKTRTIKNK HAEPSKKKAK KAGDGGVKKT KDRAGSKSKA KVPAIEVKGK PNLGQDKKKQ KRVYSEKELG IPQLNTVTPV GVTKPKGKK KGKVFVDDRE SMNTILAMVE AEKQGQIESK IMRARQLEEI REARRIEAEK KEAERKALLE NTKEQLRKKR K KNKKGGDQ ...String: MGKTRTIKNK HAEPSKKKAK KAGDGGVKKT KDRAGSKSKA KVPAIEVKGK PNLGQDKKKQ KRVYSEKELG IPQLNTVTPV GVTKPKGKK KGKVFVDDRE SMNTILAMVE AEKQGQIESK IMRARQLEEI REARRIEAEK KEAERKALLE NTKEQLRKKR K KNKKGGDQ KSSEDEGPSL KELTSTGSKA VKSKKKKKVS FATPE UniProtKB: 60S ribosomal subunit-like protein |
+Macromolecule #27: Putative NOC2 family protein
| Macromolecule | Name: Putative NOC2 family protein / type: protein_or_peptide / ID: 27 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 89.766227 KDa |
| Sequence | String: MGTAKKNLKA TKKFEKKHLK GVLERRNKVK KIKQRQQLKE KEKAKRALDD EFYKGPNGDA DKKKPGPNKA AEMSVDDFFK GGFEILSES KKKEKPAKLG KRKRDPEAEA EEQEEKSEFS ENESDVSESD ITDNDPISES ESESEGEESE DEDLGMSKSA M DALAEKDP ...String: MGTAKKNLKA TKKFEKKHLK GVLERRNKVK KIKQRQQLKE KEKAKRALDD EFYKGPNGDA DKKKPGPNKA AEMSVDDFFK GGFEILSES KKKEKPAKLG KRKRDPEAEA EEQEEKSEFS ENESDVSESD ITDNDPISES ESESEGEESE DEDLGMSKSA M DALAEKDP DFYKFLKEHD PEALDFDENA DLAEIDELSG SEDEEQPKKK QKTEEGGEGE EKKKKKKKKG KEGKDDRELT RE MVAKWKK SMEETHSLRA ARQVVIAFRC AATLHEIDED NPPRYSITSP EVFHDIVVTA LKHIPEVLQH HVPVKESSSG KTY VQTEGK KFKTLSMLIK NFTAAIIRLL STLSDDGTLK LTLSALHPLL PYLLSFRKLL KMLIKTVVAF WSQSASTDST RITA FLVIR RLVVIGDKAV RETVLKASYQ GLVQGCRVTN ANTLSGINLM KNSAAELWGL DQNLGYTTAF TSIRQLAIHL RNSII NNKN QAYRNVYNWQ YVHSLDFWSC VLSEHCSPLK EAEAGKESPL RPLIYPLVQV TLGAMRLIPT AIYFPLRFHL IRSLLR LSR ATDTYIPLAS ALLEVLQSAE MKKPPKSSTL KPLDFATAYK TPKSYLRTRV YQDGVGEQVV ELLSEFFVLW SRNIAFP EF ALPTIVALKR WMKEMRKPGK GNKNAKLGSS LVVLVQKLEM NAKFIEERRA KVDFAPKDRA QVDAFLKDLE WEKTPLGA Y VVAQRKLREE RKRLMEEARR EEERKRREEK AAEREEGSEE EEEDEEMEDA EGGEGSDEDE DEKGGGE UniProtKB: Putative NOC2 family protein |
+Macromolecule #28: rRNA-processing protein
| Macromolecule | Name: rRNA-processing protein / type: protein_or_peptide / ID: 28 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 14.710419 KDa |
| Sequence | String: MNMSETQVNI TAAPAPATTK KNLGMRKNGQ VWLPAKKAFR PTKGLTSWEL RVKKRQEQAA MKAKEREMKE EKEAERQKRI QAIKERRKK KEEKERYEQL AAKMHKKRLE RLKRKEKRNK LLNS UniProtKB: rRNA-processing protein |
+Macromolecule #29: 60S ribosomal protein L3-like protein
| Macromolecule | Name: 60S ribosomal protein L3-like protein / type: protein_or_peptide / ID: 29 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 44.456855 KDa |
| Sequence | String: MSHRKYEAPR HGSLAFLPRK RAARHRGRVK SFPKDDPKKP VHLTAAMGYK AGMTTIVRDL DRPGAKAHKK EVVEAVTIID CPPMVVVGL VGYIETPRGL RSLTTVWAEH LSDEVKRRFY KNWYKSKKKA FTKYAKKYAE NNGASITREL ERIKKYCTVV R VLAHTQIR ...String: MSHRKYEAPR HGSLAFLPRK RAARHRGRVK SFPKDDPKKP VHLTAAMGYK AGMTTIVRDL DRPGAKAHKK EVVEAVTIID CPPMVVVGL VGYIETPRGL RSLTTVWAEH LSDEVKRRFY KNWYKSKKKA FTKYAKKYAE NNGASITREL ERIKKYCTVV R VLAHTQIR KTPLKQKKAH LMEIQINGGS VADKVEFGRS LFEKPVTIDT IFEKDEMIDV IAVTKGHGFV GVTARWGTKK LP RKTHKGL RKVACIGAWH PSHVQWTVAR AGQMGYHHRT SVNHKIYRIG KGDDEANAST ETDLTKKKIT PMGGFVRYGE VNN DYVMIK GSVPGVKKRI MTLRKSLFTH TSRKALEKVE LKWIDTSSEF GHGAFQTAAE KKQFMGTLKK DLQTSA UniProtKB: Large ribosomal subunit protein uL3 |
+Macromolecule #30: 60S ribosomal protein L4-like protein
| Macromolecule | Name: 60S ribosomal protein L4-like protein / type: protein_or_peptide / ID: 30 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 39.439938 KDa |
| Sequence | String: MASRPTVTVF GADGKPTGAT EVLPKVFSAP IRPDIVKHVH TGMAKNKRQP YAVSEKAGHQ TSAESWGTGR AVARIPRVSG GGTHRAGQG AFGNMCRSGR MFAPTKIWRK WHVKINQGQK RFATASALAA SAVAPLLMAR GHQVSTVPEV PLVVDSAAVA G DAVAKTAA ...String: MASRPTVTVF GADGKPTGAT EVLPKVFSAP IRPDIVKHVH TGMAKNKRQP YAVSEKAGHQ TSAESWGTGR AVARIPRVSG GGTHRAGQG AFGNMCRSGR MFAPTKIWRK WHVKINQGQK RFATASALAA SAVAPLLMAR GHQVSTVPEV PLVVDSAAVA G DAVAKTAA AYKLLKAIGA GPDVEKVKKS KKLRAGKGKM RGRRHRQRRG PLIVYSPEHD GKELVKGFRN IPGVETCPVD AL NLLQLAP GGHLGRFIVW TSAAIKQLDA VYESKKGFFL PANIVSQADL SRLINSTEIQ SVLRAPKGEA RTKRACVQKK NPL RNKQIM LRLNPYASTF AKEKLGEVKA EEGKPPKVPA SFKELLHEA UniProtKB: 60S ribosomal protein L4-like protein |
+Macromolecule #31: 60S ribosomal protein L6
| Macromolecule | Name: 60S ribosomal protein L6 / type: protein_or_peptide / ID: 31 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 22.132994 KDa |
| Sequence | String: MSAAPTTKTF GKGTRTVPAP SEKAQKWYPA EDEAQPKKVR KAVRPWTPRK SLQPGTVLIL LAGRFRGKRV VLLKCLDQGV LLVTGPFKI NGVPLRRVNA RYVIATSVKV DLTGVDQAKI DEVAQPKYFT AEKAKEKASE EAFFKQGEKP QKKPVSSTRA A DQKAIDKA ...String: MSAAPTTKTF GKGTRTVPAP SEKAQKWYPA EDEAQPKKVR KAVRPWTPRK SLQPGTVLIL LAGRFRGKRV VLLKCLDQGV LLVTGPFKI NGVPLRRVNA RYVIATSVKV DLTGVDQAKI DEVAQPKYFT AEKAKEKASE EAFFKQGEKP QKKPVSSTRA A DQKAIDKA LIANIKKVDM LASYLASSFS LRKGDKPHLM KF UniProtKB: 60S ribosomal protein L6 |
+Macromolecule #32: 60S ribosomal protein L8
| Macromolecule | Name: 60S ribosomal protein L8 / type: protein_or_peptide / ID: 32 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 29.74449 KDa |
| Sequence | String: MPPKSGKKVA PAPFPQGKAG AKKAPKNPLL EKRPRNFGIG QDIQPKRNLS RMVKWPEYIR LQRQKKILRM RLKVPPAIAQ FQYTLDKNL AAQAFKLLNK YRPETKQEKK ERLLREATAI KEGKKKEDVS KKPYTVKYGL NHVVGLIENK KASLVLIAND V DPIELVVF ...String: MPPKSGKKVA PAPFPQGKAG AKKAPKNPLL EKRPRNFGIG QDIQPKRNLS RMVKWPEYIR LQRQKKILRM RLKVPPAIAQ FQYTLDKNL AAQAFKLLNK YRPETKQEKK ERLLREATAI KEGKKKEDVS KKPYTVKYGL NHVVGLIENK KASLVLIAND V DPIELVVF LPALCRKMGI PYAIIKGKAR LGTLVHKKTA AVVAITEVRS EDKNELAKLI SAVKEGYLEK VEDTRKRWGG GI MGFKAQK REEKRKKSLE TAIKV UniProtKB: 60S ribosomal protein L8 |
+Macromolecule #33: 60S ribosomal protein l9-like protein
| Macromolecule | Name: 60S ribosomal protein l9-like protein / type: protein_or_peptide / ID: 33 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 21.582133 KDa |
| Sequence | String: MRYIHSEETI PVPENLKVSI KSRLVTVEGP RGKLQKDLSH IAVNFSVVKK GVIGLEIHHG SRKDVAALRT VRTIINNMII GVTKGFKYK MRYVYAHFPI NVNVEKNAET GCYEVEIRNF IGEKIVRKVV MAPGVEVEIS KAQKDELILS GNSLEAVSQS A ADIQQICK ...String: MRYIHSEETI PVPENLKVSI KSRLVTVEGP RGKLQKDLSH IAVNFSVVKK GVIGLEIHHG SRKDVAALRT VRTIINNMII GVTKGFKYK MRYVYAHFPI NVNVEKNAET GCYEVEIRNF IGEKIVRKVV MAPGVEVEIS KAQKDELILS GNSLEAVSQS A ADIQQICK VRNKDIRKFL DGIYVSEKGN IVED UniProtKB: 60S ribosomal protein l9-like protein |
+Macromolecule #34: 60S ribosomal protein L12-like protein
| Macromolecule | Name: 60S ribosomal protein L12-like protein / type: protein_or_peptide / ID: 34 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 17.802676 KDa |
| Sequence | String: MPPKFDPNEI KIIHLRATGG EVGASSALAP KIGPLGLSPK KVGEDIAKAT ADWKGLRVTV RLTIQNRQAQ VSVVPTASAL IIRALKEPP RDRKKEKNIK HNKSVSLDEI IEIARTMRFK SFSKDLKGGV KEILGTAFSV GCQVDGKSPK AISDAIEAGE I DIPDE UniProtKB: 60S ribosomal protein L12-like protein |
+Macromolecule #35: 60S ribosomal protein L13
| Macromolecule | Name: 60S ribosomal protein L13 / type: protein_or_peptide / ID: 35 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 24.19118 KDa |
| Sequence | String: MAIKHNQQIP NNHFRKHWQR RVRCHFDQPG KKVTRRLARR AKAAALAPRP VDKLRPIVRC PTVKYNRRTR LGRGFTLEEL KAAGIPRLL APTIGIAVDH RRKNLSEESL AANVQRLKDY KARLILFPRK SNKPKKADTP KDQQTAETTT SLRTSFGVEQ P LAPGFTEI ...String: MAIKHNQQIP NNHFRKHWQR RVRCHFDQPG KKVTRRLARR AKAAALAPRP VDKLRPIVRC PTVKYNRRTR LGRGFTLEEL KAAGIPRLL APTIGIAVDH RRKNLSEESL AANVQRLKDY KARLILFPRK SNKPKKADTP KDQQTAETTT SLRTSFGVEQ P LAPGFTEI SKSEIPAGIE GGAYRALRKA RSDARLVGVR EKRAKEKAEA EANKK UniProtKB: 60S ribosomal protein L13 |
+Macromolecule #36: 60S ribosomal protein L14-like protein
| Macromolecule | Name: 60S ribosomal protein L14-like protein / type: protein_or_peptide / ID: 36 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 16.111901 KDa |
| Sequence | String: MAEINVEATS WRRVEVGRVL KLENGSLAAI VEIIDHKRVL ADGPSSDPKL ATPRGIVPLS RALLTPIVIP KLPRGARTGA VKKAWEAYG VDAKWKETNW AKKQLQQERR QSLTDFDRFK VMRLKKQRRF EERKALAKIK AAA UniProtKB: 60S ribosomal protein L14-like protein |
+Macromolecule #37: Ribosomal protein L15
| Macromolecule | Name: Ribosomal protein L15 / type: protein_or_peptide / ID: 37 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 24.307287 KDa |
| Sequence | String: MGALKYLEEL SKKKQSDVVR FLLRVRCWEY RQLNVIHRAS RPSRPDKARR LGYKAKQGYV IYRVRVRRGG RKRPVPKGAT YGKPTNQGV NQLKYQRSLR ATAEERVGRR CSNLRVLNSY WVNQDSTYKY YEVILVDPNH KAIRRDPRIN WICNPVHKHR E CRGLTSTG ...String: MGALKYLEEL SKKKQSDVVR FLLRVRCWEY RQLNVIHRAS RPSRPDKARR LGYKAKQGYV IYRVRVRRGG RKRPVPKGAT YGKPTNQGV NQLKYQRSLR ATAEERVGRR CSNLRVLNSY WVNQDSTYKY YEVILVDPNH KAIRRDPRIN WICNPVHKHR E CRGLTSTG KKSRGLNKGH RFNKTRAGRR KTWKRHNTLS LWRYR UniProtKB: Ribosomal protein L15 |
+Macromolecule #38: 60S ribosomal protein L16-like protein
| Macromolecule | Name: 60S ribosomal protein L16-like protein / type: protein_or_peptide / ID: 38 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 22.993195 KDa |
| Sequence | String: MSSFESVVVI DGKGHLLGRL ASIVAKQLLS GQKIVVVRCE ALNISGEFFR AKLKYHAYLR KMTRYNPTRG GPFHFRAPSR IFYKAVRGM IPHKTARGAA ALERLKVFEG VPPPYDKKKK MVVPQALRVL RLQPGRKYCT VGRLSHEVGW KYQDVVARLE E RRKAKGAA ...String: MSSFESVVVI DGKGHLLGRL ASIVAKQLLS GQKIVVVRCE ALNISGEFFR AKLKYHAYLR KMTRYNPTRG GPFHFRAPSR IFYKAVRGM IPHKTARGAA ALERLKVFEG VPPPYDKKKK MVVPQALRVL RLQPGRKYCT VGRLSHEVGW KYQDVVARLE E RRKAKGAA YYERKKLAAR QLSEAKKAAA AKVDPKVSEA LAAYGY UniProtKB: 60S ribosomal protein L16-like protein |
+Macromolecule #39: 60S ribosomal protein l17-like protein
| Macromolecule | Name: 60S ribosomal protein l17-like protein / type: protein_or_peptide / ID: 39 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 21.072207 KDa |
| Sequence | String: MVRYAATEIA PTKSARARGS YLRVSFKNTR ETAQAINGWK LQRALTFLQN VIDKKEAVPM RRYAGSTGRT AQGKQWGVSR ARWPVKSAQ FLIGLLKNAE ANADAKGLDT GNLIIKHIQV NQAPKQRRRT YRAHGRINPY MSNPCHIELI LTEAEETVQK S EAVVRDVE SHLSSRQRGV RIRRALTAA UniProtKB: Large ribosomal subunit protein uL22 |
+Macromolecule #40: Ribosomal protein L18-like protein
| Macromolecule | Name: Ribosomal protein L18-like protein / type: protein_or_peptide / ID: 40 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 24.195535 KDa |
| Sequence | String: MDLVCAFIGP LFAPGARRLN LEEIAAESSS GIDLRWKHVR SSHRKAPKSD NVYLKLLVKL YRFLARRTEA PFNKVVLRRL FMSRINRPP VSLSRIARNL KNGNEKKTVV VVATVTDDNR LLQVPKMEVA ALRFTAKARA RIEAAGGRTL TLDQLALEKP T GANTLLLR ...String: MDLVCAFIGP LFAPGARRLN LEEIAAESSS GIDLRWKHVR SSHRKAPKSD NVYLKLLVKL YRFLARRTEA PFNKVVLRRL FMSRINRPP VSLSRIARNL KNGNEKKTVV VVATVTDDNR LLQVPKMEVA ALRFTAKARA RIEAAGGRTL TLDQLALEKP T GANTLLLR GPKNAREAVK HFGFGPHKHK KPYVRSKGRK FERARGRRRS RGFKV UniProtKB: Ribosomal protein L18-like protein |
+Macromolecule #41: Ribosomal protein L19
| Macromolecule | Name: Ribosomal protein L19 / type: protein_or_peptide / ID: 41 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 317.092281 KDa |
| Sequence | String: MSEPVANDAA PGSFDLEQAR TALTSSSTTV RIAQLRAIDE KISHNSLDRP ALLGLLKVLF WTHTFYADRP SRRAVQKCLT SITKCSEAE SFVPPLVAAI RQEAQKQAIA QANSFALVEW CSLLIQNLAG TSLWEKCSKD LVLATADALE KSLQPTARGS V GHSALVIT ...String: MSEPVANDAA PGSFDLEQAR TALTSSSTTV RIAQLRAIDE KISHNSLDRP ALLGLLKVLF WTHTFYADRP SRRAVQKCLT SITKCSEAE SFVPPLVAAI RQEAQKQAIA QANSFALVEW CSLLIQNLAG TSLWEKCSKD LVLATADALE KSLQPTARGS V GHSALVIT RRGFRKLVQA DQAAVSTAIG VLAAKDAKPT AKNSVLLGVI AGVCARQPEA KAVVEDLKGQ YFTFYAREII GS RTALPSH IADGLHDFFA AFVSLEDLDK EVFPAIEKGL LRAPEVVLND LITPLVKALP KLDLSKALNS RFVKPLLSNI KSS NAVIRS GAVRAFREIA TVCRDFADLE KTADEVLGPL KTNKLASADH RVLHSEMLSG LPLSASIATK IATGLPAVTS KEAN EAALS AETLALNASA LFLLRTPEVP KALLDAYVKG LGDKKVSSRR IWILRTGDLL QAFAQEAELP AGFVAFAEAV VPSLL TIFN EVVANPTAAV QSGLVTGALV VAGLSGLLQR LENANIQALL KKAAVQKNSL ILDPKPSFVL SQRLYSKYPE DDLKWF CRA LSAIVEGLLS SSDAIKLAWT QAFIFLICSA TTPSAVRRQA IDALSDLSAR SLKPGCQFVA TEIIKGLWHW IATSEAA EK ESAAVLAKSG TSNLHLVLKA ICLSPAELNR RSGGDVDKAQ LQAQMCSLLV LAKQQLIPRA SWIDLCLKVQ VDPGQLAK D HEDALIDEIV RCTGHEQKSE ATKQAAYSAA AELVFVSPEN MTSRIVALIQ DDLDAAVVST VGPLEAAIFR TPEGTTFVD VLAKRQNVVP NKNVKDYDLL KWEEELRQQL AEKKGIQKKL TAEEQAKVSA QLKKEAEIRE NVRKIAAKLL RGFGVVKALA TGPPTDATR WMGAAVSSTL AAINAGATRI TGETGPLAFV SLSEQITSRI GSFIRPFMGV ATLRAYEVTA LPENLTEEPF E ELITRVLY RLRFVGEQRP LDTVSLIYVL PLVLYVLEKG GFAKDADEKD AQLVLAIEIL SFHTDVAFDE ALPRAEILSS LI SSMQKYS QHYKILKDCF SDMVRCIAPN ISPAEIGVLA RGAIVPQTSV RTAVLQAISA DVDMSDVNAS EEIWLACHDD IDE NAELGR EIWEESEWKT SEELGHKMIP YLESKDVQLR RAAAKSLAEV AGQHPDVVAP ILEKLRESYV ELAKPRVQQL DEFG MPKKM DLSDPWEARH GIALAFKGLA PHLEKRQLEP YFNFLIEQGP LGDQSAGVRA EMLEAANMTI EIHGKEILDR LMKTF EKVL EAPDKNSEAA DRVNEAVIIM YGALARHLKP GDKKIPVVIE RLLATLSTPS EAVQYAIAEC LPPLVRTCAD KSSKYF DEM MEILLTSKKY SEQRGAAYGL AGLVLGRGIN VLKEYRIMTQ LNSALENKKE IRQRESAMIA YELLSTILGR LFEPYVI QI VPQLLAGFGD GNADVREAAL AAAKACFAKL SSYGVKQILP TLLNGLDDDQ WRSKKGACDL LGAMAYLDPQ QLAQNLPE I IPPLTAVLND SHKEVRAAAN RSLKRFGEVI TNPEIKSLID ILLKALSDPT KYTDDALDAL IKVQFVHYLD APSLALVSR ILQRGLGDRS NTKRKASQVI GSLAHLTERK DLIAHLPVLV AGLKVAVVDP VPTTRATASR ALGSLVEKLG EDALPDLIPN LMQTLKSDT SAGDRLGSAQ ALSEVLAGLG TSRLEETLPT ILQNVESPKP AVREGFMSLF IFLPVCFGNS FANYLGKIIP P ILSGLADD VESIRETALR AGRLLVKNFA VRAVDLLLPE LERGLADDNY RIRLSSVELV GDLLFNLAGV KASANKEEDE AD QDITKEA GASLREVLGE EKRNKILSAL YVCRCDTAGA VRAAAVTVWK QLVHSPRTLK ELVPTLTQLL IKRLGSSNME HKV IASNAL GELIKKAGDG VLATLLPTLE EGLQTSRDVN AKQGICLALK ELISSASPEA LEDHEKTLIS VVRTALTDSD SDVR EAAAE AFDSLQQIIG KRAIDQVLPF LLNLLRSEEE ANNALAALLT LLTEPTRANI ILPNLIPTLI TPPISAFNAK ALASL SKVA GAAMNRRLPN IINSLMDNIV GCADETLREE LDTSFDTVIL SIDDTDGLNV VMNVLLQLIK HEDHRKRAAT GRHLAK FFS AATVNYSRYN QDIIRALLIS FDDKDMEVVK ASWNALNEFT KRLKKEEMEG LVISTRQTLL QVGVAGRELA GFELPKG IN AILPIFLQGL MNGTAEQRVA AALGISDIVD RTTEASLKPF VTQITGPLIR VVSERSTEVR SAILLTLNHL LEKMPTAL K PFLPQLQRTF AKSLADTSSE ILRSRAARAL GTLIKYTPRV DPLIAELVTG SKTTDAGVKT AMLKALYEVI SKAGSNMGE SSRAAVLGLI DMEPDENDKA MTITNAKLFG ALMKNVPADM AVGLLKNRVM VREFTTSSVL ALNAVLLESP SSLLDSPLAE DLPELLCQG IESKDAFIAD NFIMATGKYL LHDSPKAFET TKAIFTSLAK LIPPGNPGDS RRLSLVLVRT MARKQPDMVR P HLSLLAPP IFSSVRDMVI PVKLAAEAAF VQLFAVADEE SKVFDKWITA ATDLAPNVKR SMQDYFKRVT LRLGAQARER RE AEGGAGT LGLSADEVED EKELMAVGKP RVSWNRNPEK LKKPRAQSEV DPGPVPLKTV ARARQKSFKM VNLRTQKRLA ASV LGCGEG KVWLDPNEVS EISNANSRQS IRKLVADGLI IKKPVTMHSR SRARELNLAR RIGRHRGFGK RKGTADARMP QQVL WMRRQ RVLRRLLVKY RASGKIDKHL YHELYHLAKG NTFKHKRALV EHIHRAKAEK ARERQIKEEM DAKRARTKAA RERKQ ERQA AKRNALLGEE EESK UniProtKB: Ribosomal protein L19 |
+Macromolecule #42: 60S ribosomal protein L20
| Macromolecule | Name: 60S ribosomal protein L20 / type: protein_or_peptide / ID: 42 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 20.341893 KDa |
| Sequence | String: MGRLQEYQVI GRHLPTEANP NPALYRMRIF APNEVVAKSR FWYFLRGLKK VKKATGEIVS VNVISEKHPL KVKNFGIWLR YDSRSGTHN MYKEYRETSR VAAVEAMYAD MAARHRARFR SIHILKVVEL EKTEDVKRPY IKQLVAKNLS FPLPHRVPKI S TKKIFSAK RPSTFS UniProtKB: 60S ribosomal protein L20 |
+Macromolecule #43: 60S ribosomal protein l21-like protein
| Macromolecule | Name: 60S ribosomal protein l21-like protein / type: protein_or_peptide / ID: 43 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 18.159227 KDa |
| Sequence | String: MGHAAGLRSG TRYAFSRGFR KHGQIPLSTY LRTYRVGDIV DIKVNGAVQK GMPHKFYHGK TGVVYNVTKS AVGVIVYKRV KHRYIEKRI NVRIEHVKPS RSREDFLRRV KENAELKKKA KAEGVPVQLK RQPAMPREAH TVSIADNKPV TLAPVAYETT I UniProtKB: 60S ribosomal protein l21-like protein |
+Macromolecule #44: 60S ribosomal protein L22-like protein
| Macromolecule | Name: 60S ribosomal protein L22-like protein / type: protein_or_peptide / ID: 44 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 14.302563 KDa |
| Sequence | String: MAPVAKKSGA KGKGPKVTKK FIINASQPAS DKIFDVSAFE KFLNERIKVD GRVGNLGDVI KISQIGDGKV EIIAHNDLSG RYLKYLTKK FLKKMQLRDW LRVVATSKGV YELKFYNIVN DEAEEEED UniProtKB: 60S ribosomal protein L22-like protein |
+Macromolecule #45: 60S ribosomal protein l23-like protein
| Macromolecule | Name: 60S ribosomal protein l23-like protein / type: protein_or_peptide / ID: 45 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 14.674428 KDa |
| Sequence | String: MAKQKRGAPG GKLKMTLGLP VGAIMNCADN SGARNLYIIA VKGAGARLNR LPAGGVGDMV MATVKKGKPE LRKKVHPAVI VRQAKPWKR FDGVFLYFED NAGVIVNPKG EMKGSAITGP VGKEAAELWP RIASNAGVVM UniProtKB: 60S ribosomal protein l23-like protein |
+Macromolecule #46: 60S ribosomal protein L25-like protein
| Macromolecule | Name: 60S ribosomal protein L25-like protein / type: protein_or_peptide / ID: 46 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 17.175334 KDa |
| Sequence | String: MAPKDVKKGG ASKAAKGAQA KKAAQAALKG VHSHKKTKVR KSTTFHRPKT LVLSRAPKYP RKSIPHEPRL DEHKIIIHPL NTEGALKKI EEQNTLVFIV DVKANKAQIK QALKKLYDID TVKINTLIRP DGTKKAFARL TPDVDALDIA ATKLGLV UniProtKB: 60S ribosomal protein L25-like protein |
+Macromolecule #47: 60S ribosomal protein L26-like protein
| Macromolecule | Name: 60S ribosomal protein L26-like protein / type: protein_or_peptide / ID: 47 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 15.565322 KDa |
| Sequence | String: MKVRPTVSSS RRKARKAHFS APSSVRRVIM SAPLSKELRE KYNVRSIPIR KGDEVQIVRG AHKDKEGKVT SVYRLKYVIH VERVTREKA TGQTVPIGIH PSNVVITKLH LDKDRENILA RIKAGREQVA KAKGKKTAA UniProtKB: 60S ribosomal protein L26-like protein |
+Macromolecule #48: 60S ribosomal protein L27
| Macromolecule | Name: 60S ribosomal protein L27 / type: protein_or_peptide / ID: 48 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 15.803652 KDa |
| Sequence | String: MKFLKTSRVC LVTRGRYAGK KVVIIQPVDQ GTKSHPYGHA LVAGIERYPS KITRRMSKAR IEKRSKIKPF IKVINYNHLM PTRYTLELE GLKAVINADT FKEPSQREEA KKTVKKVLED RYNSGKNRWF FTPLRF UniProtKB: 60S ribosomal protein L27 |
+Macromolecule #49: 60S ribosomal protein l30-like protein
| Macromolecule | Name: 60S ribosomal protein l30-like protein / type: protein_or_peptide / ID: 49 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 11.602616 KDa |
| Sequence | String: MAPKKSKSEA QSIGAKLALV IKSGKVVLGY RSTLKALRSG KAKLILIAAN TPPLRKSELE YYSMMSKTPV HHYSGTNIEL GTACGKLFR CSTMAILDAG DSDILADQQ UniProtKB: 60S ribosomal protein l30-like protein |
+Macromolecule #50: Putative 60S ribosomal protein
| Macromolecule | Name: Putative 60S ribosomal protein / type: protein_or_peptide / ID: 50 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 13.872177 KDa |
| Sequence | String: MSSTQKKQRS AIADVVAREY TIHLHKRLHG VTFKKRAPRA IKEIKKFAQK AMGTSDVRLD PQLNKKVWEQ GIKGVPFRIR VRISRRRND EEGAKEKLYS YVQAVNVKNP KGLLTSVVEE E UniProtKB: Putative 60S ribosomal protein |
+Macromolecule #51: 60S ribosomal protein L32-like protein
| Macromolecule | Name: 60S ribosomal protein L32-like protein / type: protein_or_peptide / ID: 51 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 15.095985 KDa |
| Sequence | String: MVAAKKHVPI VKKRTKRFMR HQSDRFKCLD SAWRKPKGID NRVRRRFKGN LAMPSIGYGS NKKTKHMMPS GHKAFLVHNV KDVELLLMH NRTYAAEIAH NVSSRKRIDI ITRAKQLGVK VTNAKAKVTT EV UniProtKB: 60S ribosomal protein L32-like protein |
+Macromolecule #52: 60S ribosomal protein l33-like protein
| Macromolecule | Name: 60S ribosomal protein l33-like protein / type: protein_or_peptide / ID: 52 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 12.330337 KDa |
| Sequence | String: MPSEAGHRLY VKGRHLSYQR GRRNTHPKTS LIKIEGVDDT AAANFYLGKR VAYVYRAQKE VRGTKIRVIW GKITRPHGNS GVVRAKFTH PLPARSFGAS VRIMLYPSSI UniProtKB: 60S ribosomal protein l33-like protein |
+Macromolecule #53: Ribosomal protein l34-like protein
| Macromolecule | Name: Ribosomal protein l34-like protein / type: protein_or_peptide / ID: 53 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 13.492993 KDa |
| Sequence | String: MAPTRVTYRR RNPYNTTSNR TRVIKTPGGQ LRVLHIKKRG TAPKCGDCGI KLPGIPALRP REYATISKPK KTVQRAYGGS RCGNCVRDR IIRAFLIEEQ KIVKKVLKEQ SQAEKKASKK UniProtKB: Ribosomal protein l34-like protein |
+Macromolecule #54: dolichyl-diphosphooligosaccharide--protein glycotransferase
| Macromolecule | Name: dolichyl-diphosphooligosaccharide--protein glycotransferase type: protein_or_peptide / ID: 54 / Number of copies: 1 / Enantiomer: LEVO EC number: dolichyl-diphosphooligosaccharide-protein glycotransferase |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 105.169227 KDa |
| Sequence | String: MSSNGKVKAG QLWSKNKEEL TKILGELKTE LSQLRIQKIS SSGAKLNKIH DLRKSIARVL TVINAKQRAQ LRLFYKNKKY LPLDLRPKL TRALRRRLSK EDASRVLEKT KKRLTHFPQR KYAVKAATYS EGPALSIYHS HQREARCSLV QCCTFEPELR E SLLLLPVA ...String: MSSNGKVKAG QLWSKNKEEL TKILGELKTE LSQLRIQKIS SSGAKLNKIH DLRKSIARVL TVINAKQRAQ LRLFYKNKKY LPLDLRPKL TRALRRRLSK EDASRVLEKT KKRLTHFPQR KYAVKAATYS EGPALSIYHS HQREARCSLV QCCTFEPELR E SLLLLPVA ASLKGPNFTR RERQHRNPSE ATMSAAEPLK LLASAGKGKS TRSVLRVAIL VLIAGAAVAS RLFSVIRFES II HEFDPWF NFRATKYLVA NGFYKFWDWF DDRTWHPLGR VTGGTLYPGL MVTSGVIYHL LRFLTVPVDI RNICVLLAPG FSG LTAIAA YLLTNEMTTS PSAGLLAAAF MGIAPGYISR SVAGSYDNEA IAIFLLVFTF FLWIKALKQG SMLWGALCAL FYGY MVASW GGYAFITCLL PLHSFVLICM GRYSTRLYVA YTTWYALGTL ASMQIPFVGF LPVKTSEHMP ALGIFGFLQL LAFLD YVRS TISSRQFQTF LWLFAGGIFG LGLLGLVIAT SAGLIAPWSG RFYSLWDTGY AKIHIPIIAS VSEHQPTAWP AFFFDL NML VWLFPVGVYL CFQQLGDEHV FIIVYALFGS YFAGVMVRLM LTLTPVVCVA AAIAFSSLLD TYLNLKTPNP GQAQATE DA GKKKSGLKAA SKPAVGVYAL WGKTMMISGL TIYLLLFVLH CTWVTSNAYS SPSVVLASRL PDGSQHIIDD YREAYQWL R QNTREDAKIM SWWDYGYQIG GMADRPTLVD NNTWNNTHIA TVGKAMASRE EVSYPIMRQH EVDYVLVVFG GLLGYSGDD INKFLWMVRI AEGIWPDEVS ERAFFTPRGE YRVDAEATDT MKNSLMYKMC YYNYNNLFPP GQAVDRMRGV RLPEVGPTLN TLEEAFTSE NWIIRIYKVK DLDNLGRDHA SAAAFERGHK KKKATKKRGP RVLRVE UniProtKB: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3 |
+Macromolecule #55: 60S ribosomal protein L36
| Macromolecule | Name: 60S ribosomal protein L36 / type: protein_or_peptide / ID: 55 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 12.784053 KDa |
| Sequence | String: MSEDATPKAP VERTGLIRGL NKGHKTTRRV LKERPSRRKG AQSKRTQFVR SLIREVAGLA PYERRVIELL RNGKDKRARK FSKKKLGTF GRAKRKCEEL QRIIAESRRA H UniProtKB: 60S ribosomal protein L36 |
+Macromolecule #56: Ribosomal protein L37
| Macromolecule | Name: Ribosomal protein L37 / type: protein_or_peptide / ID: 56 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 10.660455 KDa |
| Sequence | String: MTKGTSSFGK RHNKTHGLCR RCGRRSLHNQ KKVCASCGYP AAKTRKYNWS EKAKRRKVTG TGRMRYLSTV PRRFKNGFRT GVPKGARGP AVSTSS UniProtKB: Ribosomal protein L37 |
+Macromolecule #57: 60S ribosomal protein L38-like protein
| Macromolecule | Name: 60S ribosomal protein L38-like protein / type: protein_or_peptide / ID: 57 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 9.521339 KDa |
| Sequence | String: MPQEVSDIKK FIEICRRKDA SAARIKKNPK TQQIKFKVRC QRFLYTLVLK DSDKAEKLKQ SLPPNLQIKD VPKRNKRKSS A UniProtKB: 60S ribosomal protein L38-like protein |
+Macromolecule #58: 60S ribosomal protein L39
| Macromolecule | Name: 60S ribosomal protein L39 / type: protein_or_peptide / ID: 58 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 6.297535 KDa |
| Sequence | String: MPSHKTFRIK QKLAKAQKQN RPIPQWIRLR TGNTIRYNAK RRHWRKTRLG L UniProtKB: Large ribosomal subunit protein eL39 |
+Macromolecule #59: Ribosomal protein
| Macromolecule | Name: Ribosomal protein / type: protein_or_peptide / ID: 59 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 24.055332 KDa |
| Sequence | String: MSKITVAGVR QNVAELLDYS NNVKKRNFLE TVELQIGLKN YDPQRDKRFS GTVKLPTVPR PNMSICILGD QHDIDRAKHG GIDAMSADD LKKLNKNKKL IKKLARKYDA FIASESLIKQ IPRLLGPGLS KAGKFPTPVS HSDDLSAKVN EVKSTVKFQL K KVLCMGVA ...String: MSKITVAGVR QNVAELLDYS NNVKKRNFLE TVELQIGLKN YDPQRDKRFS GTVKLPTVPR PNMSICILGD QHDIDRAKHG GIDAMSADD LKKLNKNKKL IKKLARKYDA FIASESLIKQ IPRLLGPGLS KAGKFPTPVS HSDDLSAKVN EVKSTVKFQL K KVLCMGVA VGNVGMTQEQ LVANIMLAIN YLVSLLKKGW QNVGSLTIKA TMSPPKRLY UniProtKB: Ribosomal protein |
+Macromolecule #60: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 60 / Number of copies: 1 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
+Macromolecule #61: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 61 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
+Macromolecule #62: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 62 / Number of copies: 1 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
+Macromolecule #63: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 63 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 44.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
+ About Yorodumi
About Yorodumi
-News
-Feb 9, 2022. New format data for meta-information of EMDB entries
New format data for meta-information of EMDB entries
- Version 3 of the EMDB header file is now the official format.
- The previous official version 1.9 will be removed from the archive.
Related info.: EMDB header
EMDB header
External links: wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
-Aug 12, 2020. Covid-19 info
Covid-19 info

URL: https://pdbj.org/emnavi/covid19.php
New page: Covid-19 featured information page in EM Navigator.
Related info.: Covid-19 info /
Covid-19 info /  Mar 5, 2020. Novel coronavirus structure data
Mar 5, 2020. Novel coronavirus structure data
+Mar 5, 2020. Novel coronavirus structure data
Novel coronavirus structure data
- International Committee on Taxonomy of Viruses (ICTV) defined the short name of the 2019 coronavirus as "SARS-CoV-2".
- In the structure databanks used in Yorodumi, some data are registered as the other names, "COVID-19 virus" and "2019-nCoV". Here are the details of the virus and the list of structure data.
Related info.: Yorodumi Speices /
Yorodumi Speices /  Aug 12, 2020. Covid-19 info
Aug 12, 2020. Covid-19 info
External links: COVID-19 featured content - PDBj /
COVID-19 featured content - PDBj /  Molecule of the Month (242):Coronavirus Proteases
Molecule of the Month (242):Coronavirus Proteases
+Jan 31, 2019. EMDB accession codes are about to change! (news from PDBe EMDB page)
EMDB accession codes are about to change! (news from PDBe EMDB page)
- The allocation of 4 digits for EMDB accession codes will soon come to an end. Whilst these codes will remain in use, new EMDB accession codes will include an additional digit and will expand incrementally as the available range of codes is exhausted. The current 4-digit format prefixed with “EMD-” (i.e. EMD-XXXX) will advance to a 5-digit format (i.e. EMD-XXXXX), and so on. It is currently estimated that the 4-digit codes will be depleted around Spring 2019, at which point the 5-digit format will come into force.
- The EM Navigator/Yorodumi systems omit the EMD- prefix.
Related info.: Q: What is EMD? /
Q: What is EMD? /  ID/Accession-code notation in Yorodumi/EM Navigator
ID/Accession-code notation in Yorodumi/EM Navigator
External links: EMDB Accession Codes are Changing Soon! /
EMDB Accession Codes are Changing Soon! /  Contact to PDBj
Contact to PDBj
+Jul 12, 2017. Major update of PDB
Major update of PDB
- wwPDB released updated PDB data conforming to the new PDBx/mmCIF dictionary.
- This is a major update changing the version number from 4 to 5, and with Remediation, in which all the entries are updated.
- In this update, many items about electron microscopy experimental information are reorganized (e.g. em_software).
- Now, EM Navigator and Yorodumi are based on the updated data.
External links: wwPDB Remediation /
wwPDB Remediation /  Enriched Model Files Conforming to OneDep Data Standards Now Available in the PDB FTP Archive
Enriched Model Files Conforming to OneDep Data Standards Now Available in the PDB FTP Archive
-Yorodumi
Thousand views of thousand structures
- Yorodumi is a browser for structure data from EMDB, PDB, SASBDB, etc.
- This page is also the successor to EM Navigator detail page, and also detail information page/front-end page for Omokage search.
- The word "yorodu" (or yorozu) is an old Japanese word meaning "ten thousand". "mi" (miru) is to see.
Related info.: EMDB /
EMDB /  PDB /
PDB /  SASBDB /
SASBDB /  Comparison of 3 databanks /
Comparison of 3 databanks /  Yorodumi Search /
Yorodumi Search /  Aug 31, 2016. New EM Navigator & Yorodumi /
Aug 31, 2016. New EM Navigator & Yorodumi /  Yorodumi Papers /
Yorodumi Papers /  Jmol/JSmol /
Jmol/JSmol /  Function and homology information /
Function and homology information /  Changes in new EM Navigator and Yorodumi
Changes in new EM Navigator and Yorodumi
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)