+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Baseplate of DT57C bacteriophage in the full state | |||||||||||||||
 Map data Map data | Composite map for the baseplate | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Baseplate / T5 / VIRUS / VIRAL PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||||||||
| Biological species |  Escherichia phage DT57C (virus) Escherichia phage DT57C (virus) | |||||||||||||||
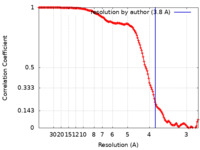
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||||||||
 Authors Authors | Ayala R / Moiseenko AV / Chen TH / Kulikov EE / Golomidova AK / Orekhov PS / Street MA / Sokolova OS / Letarov AV / Wolf M | |||||||||||||||
| Funding support |  Russian Federation, Russian Federation,  Japan, 4 items Japan, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers. Authors: Rafael Ayala / Andrey V Moiseenko / Ting-Hua Chen / Eugene E Kulikov / Alla K Golomidova / Philipp S Orekhov / Maya A Street / Olga S Sokolova / Andrey V Letarov / Matthias Wolf /     Abstract: The T5 family of viruses are tailed bacteriophages characterized by a long non-contractile tail. The bacteriophage DT57C is closely related to the paradigmal T5 phage, though it recognizes a ...The T5 family of viruses are tailed bacteriophages characterized by a long non-contractile tail. The bacteriophage DT57C is closely related to the paradigmal T5 phage, though it recognizes a different receptor (BtuB) and features highly divergent lateral tail fibers (LTF). Considerable portions of T5-like phages remain structurally uncharacterized. Here, we present the structure of DT57C determined by cryo-EM, and an atomic model of the virus, which was further explored using all-atom molecular dynamics simulations. The structure revealed a unique way of LTF attachment assisted by a dodecameric collar protein LtfC, and an unusual composition of the phage neck constructed of three protein rings. The tape measure protein (TMP) is organized within the tail tube in a three-stranded parallel α-helical coiled coil which makes direct contact with the genomic DNA. The presence of the C-terminal fragment of the TMP that remains within the tail tip suggests that the tail tip complex returns to its original state after DNA ejection. Our results provide a complete atomic structure of a T5-like phage, provide insights into the process of DNA ejection as well as a structural basis for the design of engineered phages and future mechanistic studies. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34955.map.gz emd_34955.map.gz | 221.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34955-v30.xml emd-34955-v30.xml emd-34955.xml emd-34955.xml | 29.5 KB 29.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34955_fsc.xml emd_34955_fsc.xml | 18.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_34955.png emd_34955.png | 82.1 KB | ||
| Masks |  emd_34955_msk_1.map emd_34955_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-34955.cif.gz emd-34955.cif.gz | 9.2 KB | ||
| Others |  emd_34955_half_map_1.map.gz emd_34955_half_map_1.map.gz emd_34955_half_map_2.map.gz emd_34955_half_map_2.map.gz | 221.4 MB 221.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34955 http://ftp.pdbj.org/pub/emdb/structures/EMD-34955 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34955 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34955 | HTTPS FTP |
-Validation report
| Summary document |  emd_34955_validation.pdf.gz emd_34955_validation.pdf.gz | 964.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34955_full_validation.pdf.gz emd_34955_full_validation.pdf.gz | 964 KB | Display | |
| Data in XML |  emd_34955_validation.xml.gz emd_34955_validation.xml.gz | 22.1 KB | Display | |
| Data in CIF |  emd_34955_validation.cif.gz emd_34955_validation.cif.gz | 28.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34955 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34955 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34955 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34955 | HTTPS FTP |
-Related structure data
| Related structure data |  8hqzMC  8ho3C  8hqkC  8hqoC  8hreC  8hrgC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34955.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34955.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite map for the baseplate | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.4 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_34955_msk_1.map emd_34955_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
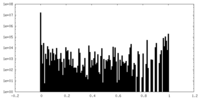
| Projections & Slices |
| ||||||||||||
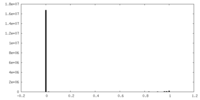
| Density Histograms |
-Half map: Half map B
| File | emd_34955_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
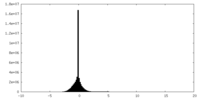
| Density Histograms |
-Half map: Half map A
| File | emd_34955_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
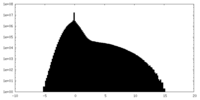
| Density Histograms |
- Sample components
Sample components
-Entire : Escherichia phage DT57C
| Entire | Name:  Escherichia phage DT57C (virus) Escherichia phage DT57C (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia phage DT57C
| Supramolecule | Name: Escherichia phage DT57C / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2681606 / Sci species name: Escherichia phage DT57C / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Baseplate hub protein
| Macromolecule | Name: Baseplate hub protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage DT57C (virus) Escherichia phage DT57C (virus) |
| Molecular weight | Theoretical: 107.114055 KDa |
| Sequence | String: MKQILDSARN YLKNNSRIKT ASLISLELPG STGTSTAFIY LTDYFRDVIY NGILYQAGKV KSISPHKQNR DLSIGSLSFT ITGTAQDEV LKLVQNGVSF LDRTVSIHQA IITEDGSILP VDPDTNGPLL YFRGRITGGG IKDNISTSGV GTSTITWNCS N QFYDFDRV ...String: MKQILDSARN YLKNNSRIKT ASLISLELPG STGTSTAFIY LTDYFRDVIY NGILYQAGKV KSISPHKQNR DLSIGSLSFT ITGTAQDEV LKLVQNGVSF LDRTVSIHQA IITEDGSILP VDPDTNGPLL YFRGRITGGG IKDNISTSGV GTSTITWNCS N QFYDFDRV NGRYTDDASH RGLEVVAGQL LPSNGAKRLE YQEDYGFFHA NKSISILAKY QVQEERYKLK SKKKLFGLSR SY SLKKYYE TVTKEVDIDF NLAAKYIPVV YGVQKIPGIP IFADTELHNP NIVYVVYAFA EGEIDGFLDF SFGDNPMICV DAN DSSART CFGTKKIVGD TMQRIASGIS SSSPSVHGQE YRYNDGNGDI RIWTYHGKSN QTASDVLVNI ARNRGFYLQN MNGN GPEYW DARYKLLDTA YAVVRFTINE NRTEIPEVSA EIQGKKVKVY HSDGRVTANS TSLNGIWQTL DYLTSDRYGA NITID QFPL QQLIQEAAIL DIIDESYQVS WQPYWRYVGW TNPLAENRQI VQMNTILDTS ESVFKNVQGL LESYGGAINN LSGQYR VTV EKYSNTPLEI NFLDTYGDLE LSDTTGRNKF NSVQASIVDP ALSWKTNSIT FYNSNYKEQD KNLDKKLQLS FANITNY YT ARSFADRELK KSRYSRTLSF SLPYQFIGIE PNDAIAFTYD RYGWDRKYFL VDEVENSREG KINVTLQEYG EDVFINSG Q VDNSGNDIPD ISNNVLPPRD FKYTPTPGGL VGSIGKNGEL SWLPSLTNNV VYYSIVHSGH AEPYIVQQLE TNPNERMIQ EIIGEPAGLA IFEIRAVDIN GRRSSPVTLS IELNSAKNLS VVSNFRVTNT ASGDVTEFVG PDVKLAWDRI PEEDIIESIF YTLEIHDPQ NRMLRSVRIE NQYTYDYLLT YNKADFALQN SDALGINRKL YFRIRAEGDD GEQSVEWASI UniProtKB: Baseplate hub protein |
-Macromolecule #2: Distal tail protein
| Macromolecule | Name: Distal tail protein / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage DT57C (virus) Escherichia phage DT57C (virus) |
| Molecular weight | Theoretical: 22.666484 KDa |
| Sequence | String: MRLPDPYTNP ELSGLGFESV NLIDNDPVIR DELPNGKVNE VKVSAQYWGI NISYPELFPD EYSVLDAFIL EYKRTGGYID VILPQYEAF RVRGNTNLVN IPAGQKGSNI TMDTQGVLTG IPKPGDLFKL SNHPKVYKIT SFNRSGNSWS INVYPDLFIT T TGAEKPVF ...String: MRLPDPYTNP ELSGLGFESV NLIDNDPVIR DELPNGKVNE VKVSAQYWGI NISYPELFPD EYSVLDAFIL EYKRTGGYID VILPQYEAF RVRGNTNLVN IPAGQKGSNI TMDTQGVLTG IPKPGDLFKL SNHPKVYKIT SFNRSGNSWS INVYPDLFIT T TGAEKPVF NGILFRTKLM NGDAFGSTLN NNGTYSGISL NLRESL UniProtKB: Distal tail protein |
-Macromolecule #3: Minor tail protein
| Macromolecule | Name: Minor tail protein / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage DT57C (virus) Escherichia phage DT57C (virus) |
| Molecular weight | Theoretical: 34.513672 KDa |
| Sequence | String: MYYSLMRESK VIVEYDGRAF HFDALSNYDV QTSYEEFKTL RRTIHRRTNY ADSVINAQTP SSISLAINFS NTLTEANFFE WLGFDRKGN TFLLPLYSNN IEPIMFNIYI VNKDNNCVYF ENCYVSTVDF SLDKNIPILN VGIESGKFSE VSTYREAASI I QGEAMSYS ...String: MYYSLMRESK VIVEYDGRAF HFDALSNYDV QTSYEEFKTL RRTIHRRTNY ADSVINAQTP SSISLAINFS NTLTEANFFE WLGFDRKGN TFLLPLYSNN IEPIMFNIYI VNKDNNCVYF ENCYVSTVDF SLDKNIPILN VGIESGKFSE VSTYREAASI I QGEAMSYS PVIVSTNGNI LPGLISASLS FQQQCSWRED KSVFDINKIY NNKRAYVNEM NASATISLYY LKRFAGDMVY NI EPETDVP LNIRNNNISI DFPLARISKR LNFSDVYKVE WDIIPTASSD PVRIDFFGEI KND UniProtKB: Minor tail protein |
-Macromolecule #4: L-shaped tail fiber assembly
| Macromolecule | Name: L-shaped tail fiber assembly / type: protein_or_peptide / ID: 4 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage DT57C (virus) Escherichia phage DT57C (virus) |
| Molecular weight | Theoretical: 15.384226 KDa |
| Sequence | String: MSTENRVIDI VVDEKVPYGL IMQFMDVDDS VYPPSEIPVN LTGYSLRGTI KSSLDENAET LASFTTSIID AAQGAAAISL SVADVTNIG EKASKERDKY NPRQRFAGYY DILMTRDVIG SEVSSFRIME GKVYISDGVT Q UniProtKB: L-shaped tail fiber assembly |
-Macromolecule #5: L-shaped tail fiber assembly
| Macromolecule | Name: L-shaped tail fiber assembly / type: protein_or_peptide / ID: 5 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage DT57C (virus) Escherichia phage DT57C (virus) |
| Molecular weight | Theoretical: 113.712461 KDa |
| Sequence | String: MALKTKIIVQ QILNIDDTTT TASKYPKYTV VLGNSISSIT AGELTAAVEA SAGSAAAAKG SEIAAKESEL NAKDSENEAA ISAGASEES ASQSAASAAE SERQAGLSKG SADNSAASAQ ESEGFRDSAE LAAQNAEQSR LLAEQAKTAA QQAQTAAEAA K TGAETAKD ...String: MALKTKIIVQ QILNIDDTTT TASKYPKYTV VLGNSISSIT AGELTAAVEA SAGSAAAAKG SEIAAKESEL NAKDSENEAA ISAGASEES ASQSAASAAE SERQAGLSKG SADNSAASAQ ESEGFRDSAE LAAQNAEQSR LLAEQAKTAA QQAQTAAEAA K TGAETAKD GADAAATTAG EHAAAARQSE LNAKISETNA AGSATEAGDK AIDATTEADR AKAEADRATQ IVDSKLDKVD IS GFIKVYK TKAEADADVV NRVLDEKVLV WNQTNSKYGW YKVAGTAETP VLELVETEQK LTSVNNVRAD DAGNVQITLP GGN PSLWLG EVTWFPYDKD SGVGYPGVLP ADGREVLRVD YPDTWEAIEA GLIPSVSEAE WQAGASLYFS TGDGSTTFRL PDMM QGQAF RAPTKGEEDA GVIKDQIPYV VTVNGISPDA ITGNVEIDTS LQGTVSINQG GTGATTKEDA RIALELYSTT EVDSA LADK ADIATTYTKT EVDSALADKA DIATTYTKVE VDSALADAKT QSDTDYLLKA NNLSDLADRA AAWLNVRPIG STPLAG DPV GDYDAVTKRW VENKINTGTV GPTMNGVMNY GVGDFHLRDS RAYIQPYEVV SDGQLLNRAD WPELWAYAQM LSPISDA DW LADPTKRGQY SLGDGSTTFR VPDRNGVQTG SISALFGRGD GGASSTGGTI LDSAAPNITG SFGRLVYAST GTIYEANT G TGAFSAVLSQ AKYKRLSEIS AADGTAATYP SGFEFFASNS SPVYGRGSTE VRPKAFTGVW VIRASGGFVA ANTSWSVIN GDATRPADGT TADGGEIISR YNVNGVREAQ MSWRIRAQIG AEHYARLNVY NATANRTAVY DFNDLGTFSA ENLHSKGAIY SDGNLTIQN QGWPGINFKS NRYNTPATQI GGSTIIEVSG TDGNVSGVNL IRRRGDGNQA GQIIVSFPTT GGAIALQGTS G IEYKKDVT DADAQEAMDR INGQRLVNFV YKDDEQERVR FGVIAEEAEL IAPQYIKHNQ VSYEDILDEE GNKIGEKTRD RP SVDVNPI VMDLMGCVQA LNAKIAALEA RIAELESKE UniProtKB: L-shaped tail fiber assembly |
-Macromolecule #6: Tape measure protein
| Macromolecule | Name: Tape measure protein / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage DT57C (virus) Escherichia phage DT57C (virus) |
| Molecular weight | Theoretical: 132.626406 KDa |
| Sequence | String: MTDKLIRELL IDVKQKGATR TAKSIENVSD ALENAAAASE LTNEQLGKMP KTLYSIERAA DRAAKSLTKM QASRGMAGIT KSIDSIGAK LDDLSIAMIE VADKLEAGFD GVSRSVKVMG NDVAAATEKV QDRLYDTNRV LGGTARGFND TAGAAGRASR A IGNTSGSA ...String: MTDKLIRELL IDVKQKGATR TAKSIENVSD ALENAAAASE LTNEQLGKMP KTLYSIERAA DRAAKSLTKM QASRGMAGIT KSIDSIGAK LDDLSIAMIE VADKLEAGFD GVSRSVKVMG NDVAAATEKV QDRLYDTNRV LGGTARGFND TAGAAGRASR A IGNTSGSA RGATRDFAAM AKVGGGLPLL YAAIASNIFV LQSAFEQLKL GDQLNRLEEF GVIVGTQTGT PVQSLARSLQ EA AGYAISF EEAMRQASSA SAYGFDAEQL NKFGLVARRA AAVLGVDMTD ALNRVIKGVS KQEIELLDEL GVTIRLNDAY ADY VKQLNA ANTGITYNIN SLTTFQKQQA YANAVIAEST KRFGYLDEVL RATPWEQFAA NADAALRTIQ QAAAKYLGPV IDAI NTVFY TSQASISAEA ARAQEQTNKQ IDPTNVGAVA LSLSASEEGY NKALDMYKES LDKRNKLKSE FDKRMEQADF YTKLA IRQV GEGIPAGLAT AGASEANKKF VEETAAMGLQ VARLDKEVTD STENLNAWKS AYQAAGAAAA KANPEFQKQI NLQRDT TDP GAVYDFNSTV LKGLTEQQKA YNQTKKTASD LANDIQNVAQ NTDTAAKTSA TLADAIKNIE SLSLGTGKSA DEYVKNL NL GYNTLSEMKT ASQALSEYVK LTGNETKNQL AVQQKIADVY NQTKDKEKAQ EAGRRLELQQ LEEQEAALRR VLQTNQGN K AVEREIEKIQ LEKIKLTNQG MEAQKKVKDY TDKILGVDRE IALLNNRTMT DTQYRLAQLN LELTIEKEKY EWYTKQADK QKEAEQSRRA QAQIEREIWK FHQDQQAEMT SKRQEAFENT LTSMFPLAGE MQKMEMQLDF YTQMKELTKD NANEQMRWNA EIAKTRAQM SALTAQRNAQ MQSSVGSSLG AVYTPTTGLS GEDKKFADMG NQLASYDQAI SKLSELNSEA TAVAQSMGNL A NAMIQFSQ GSLDTTSTIA VGMQTVASMI QYSTSQQVSA IDQAIAAEQK RDGKSEASKA KLKKLEAEKL KIQQDAAKKQ II IQTAVAV MQAATAVPYP FSIPLMIAAG LAGALALAQA SSASGMSSIG DSGADTASYL TLGERQKNID VSMSANAGEL SYV RGDKGI GNANSFVPRA EGGNMYPGVS YQMGEHGTEV VTPMVPMKAT PNDELKNSSN STAGRPIILN ISAMDAASFR EFAS SNSGA LRDAVELALN ENGASLKTLG NS UniProtKB: Tape measure protein |
-Macromolecule #7: Tail tube protein
| Macromolecule | Name: Tail tube protein / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage DT57C (virus) Escherichia phage DT57C (virus) |
| Molecular weight | Theoretical: 50.759051 KDa |
| Sequence | String: MSLQLLRNTR IFVSTVKTGH DTTNTQEILV QDDISWGQDS NSTDITVNEA GPRPTRGSKR FNDSLNAAEW SFSTYILPYE DTTTGKQIV PDYMLWHALS SGKAIDLDGD TGAHNNATNF MVNFKDNAYH ELAMLHIYIL TDNAWSYIDS CQINQAEVNV D IEDIGRVT ...String: MSLQLLRNTR IFVSTVKTGH DTTNTQEILV QDDISWGQDS NSTDITVNEA GPRPTRGSKR FNDSLNAAEW SFSTYILPYE DTTTGKQIV PDYMLWHALS SGKAIDLDGD TGAHNNATNF MVNFKDNAYH ELAMLHIYIL TDNAWSYIDS CQINQAEVNV D IEDIGRVT WSGNGNQLIP LDEQPFDPDA LGIDDETYMT IQSSYIKNKL TILKIKDMDS DKEYDIPITG GTFTINNNIT YL TPNIMSR VNIPIGSFTG AFELTGSLTA YLNDKSLGSM ELYKDLVRTL KVVNRFEIAL ILGGEYDDER PAAVLVAKQA HVN IPTIET DDVLGTSVEF KAIPTDLDTG DEGYLGFSNK YTKTTVANLI ATGDGAETPP ILVESITVKS AADATSVTNS DTLQ MSVEV TPPEATNTAV TWSISSGDAA TIDAESGLLT ADVSKTGEVI VKAVAKDGSG VEGTKTITVS AGE UniProtKB: Tail tube protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 67.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

























 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)