[English] 日本語
 Yorodumi
Yorodumi- EMDB-34973: Curved tail segment of the DT57C bacteriophage in the full state -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Curved tail segment of the DT57C bacteriophage in the full state | |||||||||||||||
 Map data Map data | Map of a curved segment of the tail | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Tail tube / T5 / VIRUS / VIRAL PROTEIN | |||||||||||||||
| Biological species |  Escherichia phage DT57C (virus) Escherichia phage DT57C (virus) | |||||||||||||||
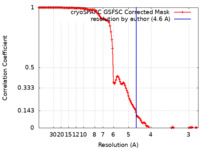
| Method | single particle reconstruction / cryo EM / Resolution: 4.6 Å | |||||||||||||||
 Authors Authors | Ayala R / Moiseenko AV / Chen TH / Kulikov EE / Golomidova AK / Orekhov PS / Street MA / Sokolova OS / Letarov AV / Wolf M | |||||||||||||||
| Funding support |  Russian Federation, Russian Federation,  Japan, 4 items Japan, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers. Authors: Rafael Ayala / Andrey V Moiseenko / Ting-Hua Chen / Eugene E Kulikov / Alla K Golomidova / Philipp S Orekhov / Maya A Street / Olga S Sokolova / Andrey V Letarov / Matthias Wolf /     Abstract: The T5 family of viruses are tailed bacteriophages characterized by a long non-contractile tail. The bacteriophage DT57C is closely related to the paradigmal T5 phage, though it recognizes a ...The T5 family of viruses are tailed bacteriophages characterized by a long non-contractile tail. The bacteriophage DT57C is closely related to the paradigmal T5 phage, though it recognizes a different receptor (BtuB) and features highly divergent lateral tail fibers (LTF). Considerable portions of T5-like phages remain structurally uncharacterized. Here, we present the structure of DT57C determined by cryo-EM, and an atomic model of the virus, which was further explored using all-atom molecular dynamics simulations. The structure revealed a unique way of LTF attachment assisted by a dodecameric collar protein LtfC, and an unusual composition of the phage neck constructed of three protein rings. The tape measure protein (TMP) is organized within the tail tube in a three-stranded parallel α-helical coiled coil which makes direct contact with the genomic DNA. The presence of the C-terminal fragment of the TMP that remains within the tail tip suggests that the tail tip complex returns to its original state after DNA ejection. Our results provide a complete atomic structure of a T5-like phage, provide insights into the process of DNA ejection as well as a structural basis for the design of engineered phages and future mechanistic studies. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34973.map.gz emd_34973.map.gz | 200.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34973-v30.xml emd-34973-v30.xml emd-34973.xml emd-34973.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34973_fsc.xml emd_34973_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_34973.png emd_34973.png | 63.5 KB | ||
| Filedesc metadata |  emd-34973.cif.gz emd-34973.cif.gz | 5.9 KB | ||
| Others |  emd_34973_half_map_1.map.gz emd_34973_half_map_1.map.gz emd_34973_half_map_2.map.gz emd_34973_half_map_2.map.gz | 195.1 MB 195.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34973 http://ftp.pdbj.org/pub/emdb/structures/EMD-34973 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34973 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34973 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34973.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34973.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of a curved segment of the tail | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.4 Å | ||||||||||||||||||||||||||||||||||||
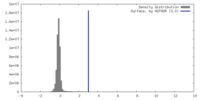
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map A
| File | emd_34973_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
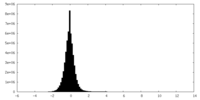
| Density Histograms |
-Half map: Half map B
| File | emd_34973_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
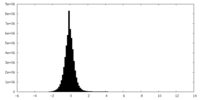
| Density Histograms |
- Sample components
Sample components
-Entire : Escherichia phage DT57C
| Entire | Name:  Escherichia phage DT57C (virus) Escherichia phage DT57C (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia phage DT57C
| Supramolecule | Name: Escherichia phage DT57C / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2681606 / Sci species name: Escherichia phage DT57C / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Tail tube protein (TTP)
| Macromolecule | Name: Tail tube protein (TTP) / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage DT57C (virus) Escherichia phage DT57C (virus) |
| Sequence | String: MSLQLLRNTR IFVSTVKTGH DTTNTQEILV QDDISWGQDS NSTDITVNEA GPRPTRGSKR FNDSLNAAE WSFSTYILPY EDTTTGKQIV PDYMLWHALS SGKAIDLDGD TGAHNNATNF M VNFKDNAY HELAMLHIYI LTDNAWSYID SCQINQAEVN VDIEDIGRVT ...String: MSLQLLRNTR IFVSTVKTGH DTTNTQEILV QDDISWGQDS NSTDITVNEA GPRPTRGSKR FNDSLNAAE WSFSTYILPY EDTTTGKQIV PDYMLWHALS SGKAIDLDGD TGAHNNATNF M VNFKDNAY HELAMLHIYI LTDNAWSYID SCQINQAEVN VDIEDIGRVT WSGNGNQLIP LD EQPFDPD ALGIDDETYM TIQSSYIKNK LTILKIKDMD SDKEYDIPIT GGTFTINNNI TYL TPNIMS RVNIPIGSFT GAFELTGSLT AYLNDKSLGS MELYKDLVRT LKVVNRFEIA LILG GEYDD ERPAAVLVAK QAHVNIPTIE TDDVLGTSVE FKAIPTDLDT GDEGYLGFSN KYTKT TVAN LIATGDGAET PPILVESITV KSAADATSVT NSDTLQMSVE VTPPEATNTA VTWSIS SGD AATIDAESGL LTADVSKTGE VIVKAVAKDG SGVEGTKTIT VSAGE |
-Macromolecule #2: Tape measure protein (TMP)
| Macromolecule | Name: Tape measure protein (TMP) / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage DT57C (virus) Escherichia phage DT57C (virus) |
| Sequence | String: MTDKLIRELL IDVKQKGATR TAKSIENVSD ALENAAAASE LTNEQLGKMP KTLYSIERAA DRAAKSLTK MQASRGMAGI TKSIDSIGAK LDDLSIAMIE VADKLEAGFD GVSRSVKVMG N DVAAATEK VQDRLYDTNR VLGGTARGFN DTAGAAGRAS RAIGNTSGSA ...String: MTDKLIRELL IDVKQKGATR TAKSIENVSD ALENAAAASE LTNEQLGKMP KTLYSIERAA DRAAKSLTK MQASRGMAGI TKSIDSIGAK LDDLSIAMIE VADKLEAGFD GVSRSVKVMG N DVAAATEK VQDRLYDTNR VLGGTARGFN DTAGAAGRAS RAIGNTSGSA RGATRDFAAM AK VGGGLPL LYAAIASNIF VLQSAFEQLK LGDQLNRLEE FGVIVGTQTG TPVQSLARSL QEA AGYAIS FEEAMRQASS ASAYGFDAEQ LNKFGLVARR AAAVLGVDMT DALNRVIKGV SKQE IELLD ELGVTIRLND AYADYVKQLN AANTGITYNI NSLTTFQKQQ AYANAVIAES TKRFG YLDE VLRATPWEQF AANADAALRT IQQAAAKYLG PVIDAINTVF YTSQASISAE AARAQE QTN KQIDPTNVGA VALSLSASEE GYNKALDMYK ESLDKRNKLK SEFDKRMEQA DFYTKLA IR QVGEGIPAGL ATAGASEANK KFVEETAAMG LQVARLDKEV TDSTENLNAW KSAYQAAG A AAAKANPEFQ KQINLQRDTT DPGAVYDFNS TVLKGLTEQQ KAYNQTKKTA SDLANDIQN VAQNTDTAAK TSATLADAIK NIESLSLGTG KSADEYVKNL NLGYNTLSEM KTASQALSEY VKLTGNETK NQLAVQQKIA DVYNQTKDKE KAQEAGRRLE LQQLEEQEAA LRRVLQTNQG N KAVEREIE KIQLEKIKLT NQGMEAQKKV KDYTDKILGV DREIALLNNR TMTDTQYRLA QL NLELTIE KEKYEWYTKQ ADKQKEAEQS RRAQAQIERE IWKFHQDQQA EMTSKRQEAF ENT LTSMFP LAGEMQKMEM QLDFYTQMKE LTKDNANEQM RWNAEIAKTR AQMSALTAQR NAQM QSSVG SSLGAVYTPT TGLSGEDKKF ADMGNQLASY DQAISKLSEL NSEATAVAQS MGNLA NAMI QFSQGSLDTT STIAVGMQTV ASMIQYSTSQ QVSAIDQAIA AEQKRDGKSE ASKAKL KKL EAEKLKIQQD AAKKQIIIQT AVAVMQAATA VPYPFSIPLM IAAGLAGALA LAQASSA SG MSSIGDSGAD TASYLTLGER QKNIDVSMSA NAGELSYVRG DKGIGNANSF VPRAEGGN M YPGVSYQMGE HGTEVVTPMV PMKATPNDEL KNSSNSTAGR PIILNISAMD AASFREFAS SNSGALRDAV ELALNENGAS LKTLGNS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 67.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)