+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human ATP synthase F1 domain, state 3b | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | MEMBRANE PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of cell adhesion involved in substrate-bound cell migration / Formation of ATP by chemiosmotic coupling / Cristae formation / ATP biosynthetic process / angiostatin binding / Mitochondrial protein import / : / : / cellular response to interleukin-7 / oxidative phosphorylation ...negative regulation of cell adhesion involved in substrate-bound cell migration / Formation of ATP by chemiosmotic coupling / Cristae formation / ATP biosynthetic process / angiostatin binding / Mitochondrial protein import / : / : / cellular response to interleukin-7 / oxidative phosphorylation / response to muscle activity / proton-transporting ATP synthase complex / : / : / mitochondrial nucleoid / negative regulation of endothelial cell proliferation / MHC class I protein binding / proton motive force-driven ATP synthesis / proton motive force-driven mitochondrial ATP synthesis / cellular response to nitric oxide / positive regulation of blood vessel endothelial cell migration / proton-transporting ATP synthase complex, catalytic core F(1) / H+-transporting two-sector ATPase / Mitochondrial protein degradation / proton-transporting ATPase activity, rotational mechanism / proton-transporting ATP synthase activity, rotational mechanism / cellular response to dexamethasone stimulus / proton transmembrane transport / generation of precursor metabolites and energy / regulation of intracellular pH / mitochondrial membrane / ADP binding / Transcriptional activation of mitochondrial biogenesis / lipid metabolic process / osteoblast differentiation / protease binding / angiogenesis / response to ethanol / mitochondrial inner membrane / mitochondrial matrix / membrane raft / cell surface / ATP hydrolysis activity / mitochondrion / RNA binding / extracellular exosome / ATP binding / membrane / nucleus / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.02 Å | |||||||||||||||
 Authors Authors | Lai Y / Zhang Y / Liu F / Gao Y / Gong H / Rao Z | |||||||||||||||
| Funding support |  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2023 Journal: Mol Cell / Year: 2023Title: Structure of the human ATP synthase. Authors: Yuezheng Lai / Yuying Zhang / Shan Zhou / Jinxu Xu / Zhanqiang Du / Ziyan Feng / Long Yu / Ziqing Zhao / Weiwei Wang / Yanting Tang / Xiuna Yang / Luke W Guddat / Fengjiang Liu / Yan Gao / ...Authors: Yuezheng Lai / Yuying Zhang / Shan Zhou / Jinxu Xu / Zhanqiang Du / Ziyan Feng / Long Yu / Ziqing Zhao / Weiwei Wang / Yanting Tang / Xiuna Yang / Luke W Guddat / Fengjiang Liu / Yan Gao / Zihe Rao / Hongri Gong /   Abstract: Biological energy currency ATP is produced by FF-ATP synthase. However, the molecular mechanism for human ATP synthase action remains unknown. Here, we present snapshot images for three main ...Biological energy currency ATP is produced by FF-ATP synthase. However, the molecular mechanism for human ATP synthase action remains unknown. Here, we present snapshot images for three main rotational states and one substate of human ATP synthase using cryoelectron microscopy. These structures reveal that the release of ADP occurs when the β subunit of FF-ATP synthase is in the open conformation, showing how ADP binding is coordinated during synthesis. The accommodation of the symmetry mismatch between F and F motors is resolved by the torsional flexing of the entire complex, especially the γ subunit, and the rotational substep of the c subunit. Water molecules are identified in the inlet and outlet half-channels, suggesting that the proton transfer in these two half-channels proceed via a Grotthus mechanism. Clinically relevant mutations are mapped to the structure, showing that they are mainly located at the subunit-subunit interfaces, thus causing instability of the complex. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34576.map.gz emd_34576.map.gz | 484.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34576-v30.xml emd-34576-v30.xml emd-34576.xml emd-34576.xml | 20 KB 20 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34576.png emd_34576.png | 119.8 KB | ||
| Filedesc metadata |  emd-34576.cif.gz emd-34576.cif.gz | 6.4 KB | ||
| Others |  emd_34576_half_map_1.map.gz emd_34576_half_map_1.map.gz emd_34576_half_map_2.map.gz emd_34576_half_map_2.map.gz | 475 MB 475 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34576 http://ftp.pdbj.org/pub/emdb/structures/EMD-34576 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34576 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34576 | HTTPS FTP |
-Validation report
| Summary document |  emd_34576_validation.pdf.gz emd_34576_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34576_full_validation.pdf.gz emd_34576_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_34576_validation.xml.gz emd_34576_validation.xml.gz | 19 KB | Display | |
| Data in CIF |  emd_34576_validation.cif.gz emd_34576_validation.cif.gz | 22.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34576 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34576 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34576 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34576 | HTTPS FTP |
-Related structure data
| Related structure data |  8h9pMC  8h9eC  8h9fC  8h9gC  8h9iC  8h9jC  8h9kC  8h9lC  8h9mC  8h9nC  8h9qC  8h9rC  8h9sC  8h9tC  8h9uC  8h9vC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34576.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34576.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.73 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34576_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
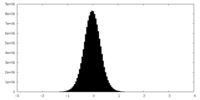
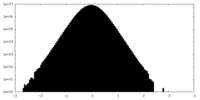
| Density Histograms |
-Half map: #1
| File | emd_34576_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
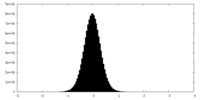
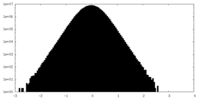
| Density Histograms |
- Sample components
Sample components
-Entire : Human ATP synthase
| Entire | Name: Human ATP synthase |
|---|---|
| Components |
|
-Supramolecule #1: Human ATP synthase
| Supramolecule | Name: Human ATP synthase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 600 KDa |
-Macromolecule #1: ATP synthase subunit alpha, mitochondrial
| Macromolecule | Name: ATP synthase subunit alpha, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 55.27616 KDa |
| Sequence | String: QKTGTAEMSS ILEERILGAD TSVDLEETGR VLSIGDGIAR VHGLRNVQAE EMVEFSSGLK GMSLNLEPDN VGVVVFGNDK LIKEGDIVK RTGAIVDVPV GEELLGRVVD ALGNAIDGKG PIGSKTRRRV GLKAPGIIPR ISVREPMQTG IKAVDSLVPI G RGQRELII ...String: QKTGTAEMSS ILEERILGAD TSVDLEETGR VLSIGDGIAR VHGLRNVQAE EMVEFSSGLK GMSLNLEPDN VGVVVFGNDK LIKEGDIVK RTGAIVDVPV GEELLGRVVD ALGNAIDGKG PIGSKTRRRV GLKAPGIIPR ISVREPMQTG IKAVDSLVPI G RGQRELII GDRQTGKTSI AIDTIINQKR FNDGSDEKKK LYCIYVAIGQ KRSTVAQLVK RLTDADAMKY TIVVSATASD AA PLQYLAP YSGCSMGEYF RDNGKHALII YDDLSKQAVA YRQMSLLLRR PPGREAYPGD VFYLHSRLLE RAAKMNDAFG GGS LTALPV IETQAGDVSA YIPTNVISIT DGQIFLETEL FYKGIRPAIN VGLSVSRVGS AAQTRAMKQV AGTMKLELAQ YREV AAFAQ FGSDLDAATQ QLLSRGVRLT ELLKQGQYSP MAIEEQVAVI YAGVRGYLDK LEPSKITKFE NAFLSHVVSQ HQALL GTIR ADGKISEQSD AKLKEIVTNF LAGFEA UniProtKB: ATP synthase subunit alpha, mitochondrial |
-Macromolecule #2: ATP synthase subunit beta, mitochondrial
| Macromolecule | Name: ATP synthase subunit beta, mitochondrial / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 51.821965 KDa |
| Sequence | String: AQTSPSPKAG AATGRIVAVI GAVVDVQFDE GLPPILNALE VQGRETRLVL EVAQHLGEST VRTIAMDGTE GLVRGQKVLD SGAPIKIPV GPETLGRIMN VIGEPIDERG PIKTKQFAPI HAEAPEFMEM SVEQEILVTG IKVVDLLAPY AKGGKIGLFG G AGVGKTVL ...String: AQTSPSPKAG AATGRIVAVI GAVVDVQFDE GLPPILNALE VQGRETRLVL EVAQHLGEST VRTIAMDGTE GLVRGQKVLD SGAPIKIPV GPETLGRIMN VIGEPIDERG PIKTKQFAPI HAEAPEFMEM SVEQEILVTG IKVVDLLAPY AKGGKIGLFG G AGVGKTVL IMELINNVAK AHGGYSVFAG VGERTREGND LYHEMIESGV INLKDATSKV ALVYGQMNEP PGARARVALT GL TVAEYFR DQEGQDVLLF IDNIFRFTQA GSEVSALLGR IPSAVGYQPT LATDMGTMQE RITTTKKGSI TSVQAIYVPA DDL TDPAPA TTFAHLDATT VLSRAIAELG IYPAVDPLDS TSRIMDPNIV GSEHYDVARG VQKILQDYKS LQDIIAILGM DELS EEDKL TVSRARKIQR FLSQPFQVAE VFTGHMGKLV PLKETIKGFQ QILAGEYDHL PEQAFYMVGP IEEAVAKADK LAEEH SS UniProtKB: ATP synthase subunit beta, mitochondrial |
-Macromolecule #3: ATP synthase subunit gamma, mitochondrial
| Macromolecule | Name: ATP synthase subunit gamma, mitochondrial / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 30.207752 KDa |
| Sequence | String: ATLKDITRRL KSIKNIQKIT KSMKMVAAAK YARAERELKP ARIYGLGSLA LYEKADIKGP EDKKKHLLIG VSSDRGLCGA IHSSIAKQM KSEVATLTAA GKEVMLVGIG DKIRGILYRT HSDQFLVAFK EVGRKPPTFG DASVIALELL NSGYEFDEGS I IFNKFRSV ...String: ATLKDITRRL KSIKNIQKIT KSMKMVAAAK YARAERELKP ARIYGLGSLA LYEKADIKGP EDKKKHLLIG VSSDRGLCGA IHSSIAKQM KSEVATLTAA GKEVMLVGIG DKIRGILYRT HSDQFLVAFK EVGRKPPTFG DASVIALELL NSGYEFDEGS I IFNKFRSV ISYKTEEKPI FSLNTVASAD SMSIYDDIDA DVLQNYQEYN LANIIYYSLK ESTTSEQSAR MTAMDNASKN AS EMIDKLT LTFNRTRQAV ITKELIEIIS GAAALD UniProtKB: ATP synthase subunit gamma, mitochondrial |
-Macromolecule #4: ATP synthase subunit O, mitochondrial
| Macromolecule | Name: ATP synthase subunit O, mitochondrial / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 20.904488 KDa |
| Sequence | String: FAKLVRPPVQ VYGIEGRYAT ALYSAASKQN KLEQVEKELL RVAQILKEPK VAASVLNPYV KRSIKVKSLN DITAKERFSP LTTNLINLL AENGRLSNTQ GVVSAFSTMM SVHRGEVPCT VTSASPLEEA TLSELKTVLK SFLSQGQVLK LEAKTDPSIL G GMIVRIGE KYVDMSVKTK IQKLGRAMRE IV UniProtKB: ATP synthase subunit O, mitochondrial |
-Macromolecule #5: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 3 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 5 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #7: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 7 / Number of copies: 2 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris X / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: AlphaFold |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.02 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 23272 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)