[English] 日本語
 Yorodumi
Yorodumi- EMDB-33501: Structure of photosynthetic LH1-RC super-complex of Rhodopila glo... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of photosynthetic LH1-RC super-complex of Rhodopila globiformis | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationorganelle inner membrane / plasma membrane-derived chromatophore membrane / plasma membrane light-harvesting complex / bacteriochlorophyll binding / photosynthesis, light reaction / photosynthetic electron transport in photosystem II / electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity / electron transfer activity / iron ion binding / heme binding ...organelle inner membrane / plasma membrane-derived chromatophore membrane / plasma membrane light-harvesting complex / bacteriochlorophyll binding / photosynthesis, light reaction / photosynthetic electron transport in photosystem II / electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity / electron transfer activity / iron ion binding / heme binding / membrane / metal ion binding / plasma membrane Similarity search - Function | |||||||||||||||||||||
| Biological species |  Rhodopila globiformis (bacteria) Rhodopila globiformis (bacteria) | |||||||||||||||||||||
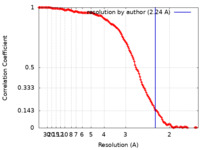
| Method | single particle reconstruction / cryo EM / Resolution: 2.24 Å | |||||||||||||||||||||
 Authors Authors | Tani K / Kanno R / Kurosawa K / Takaichi S / Nagashima KVP / Hall M / Yu L-J / Kimura Y / Madigan MT / Mizoguchi A ...Tani K / Kanno R / Kurosawa K / Takaichi S / Nagashima KVP / Hall M / Yu L-J / Kimura Y / Madigan MT / Mizoguchi A / Humbel BM / Wang-Otomo Z-Y | |||||||||||||||||||||
| Funding support |  Japan, 6 items Japan, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2022 Journal: Commun Biol / Year: 2022Title: An LH1-RC photocomplex from an extremophilic phototroph provides insight into origins of two photosynthesis proteins. Authors: Kazutoshi Tani / Ryo Kanno / Keigo Kurosawa / Shinichi Takaichi / Kenji V P Nagashima / Malgorzata Hall / Long-Jiang Yu / Yukihiro Kimura / Michael T Madigan / Akira Mizoguchi / Bruno M ...Authors: Kazutoshi Tani / Ryo Kanno / Keigo Kurosawa / Shinichi Takaichi / Kenji V P Nagashima / Malgorzata Hall / Long-Jiang Yu / Yukihiro Kimura / Michael T Madigan / Akira Mizoguchi / Bruno M Humbel / Zheng-Yu Wang-Otomo /    Abstract: Rhodopila globiformis is the most acidophilic of anaerobic purple phototrophs, growing optimally in culture at pH 5. Here we present a cryo-EM structure of the light-harvesting 1-reaction center (LH1- ...Rhodopila globiformis is the most acidophilic of anaerobic purple phototrophs, growing optimally in culture at pH 5. Here we present a cryo-EM structure of the light-harvesting 1-reaction center (LH1-RC) complex from Rhodopila globiformis at 2.24 Å resolution. All purple bacterial cytochrome (Cyt, encoded by the gene pufC) subunit-associated RCs with known structures have their N-termini truncated. By contrast, the Rhodopila globiformis RC contains a full-length tetra-heme Cyt with its N-terminus embedded in the membrane forming an α-helix as the membrane anchor. Comparison of the N-terminal regions of the Cyt with PufX polypeptides widely distributed in Rhodobacter species reveals significant structural similarities, supporting a longstanding hypothesis that PufX is phylogenetically related to the N-terminus of the RC-bound Cyt subunit and that a common ancestor of phototrophic Proteobacteria contained a full-length tetra-heme Cyt subunit that evolved independently through partial deletions of its pufC gene. Eleven copies of a novel γ-like polypeptide were also identified in the bacteriochlorophyll a-containing Rhodopila globiformis LH1 complex; γ-polypeptides have previously been found only in the LH1 of bacteriochlorophyll b-containing species. These features are discussed in relation to their predicted functions of stabilizing the LH1 structure and regulating quinone transport under the warm acidic conditions. #1:  Journal: Commun Biol / Year: 2022 Journal: Commun Biol / Year: 2022Title: An LH1-RC photocomplex from an extremophilic phototroph provides insight into origins of two photosynthesis proteins Authors: Tani K / Kanno R / Kurosawa K / Takaichi S / Nagashima KVP / Hall M / Yu L-J / Kimura Y / Madigan MT / Mizoguchi A / Humbel BM / Wang-Otomo Z-Y | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33501.map.gz emd_33501.map.gz | 229.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33501-v30.xml emd-33501-v30.xml emd-33501.xml emd-33501.xml | 29.1 KB 29.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33501_fsc.xml emd_33501_fsc.xml | 13.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_33501.png emd_33501.png | 207.5 KB | ||
| Others |  emd_33501_half_map_1.map.gz emd_33501_half_map_1.map.gz emd_33501_half_map_2.map.gz emd_33501_half_map_2.map.gz | 194 MB 193.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33501 http://ftp.pdbj.org/pub/emdb/structures/EMD-33501 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33501 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33501 | HTTPS FTP |
-Validation report
| Summary document |  emd_33501_validation.pdf.gz emd_33501_validation.pdf.gz | 846.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33501_full_validation.pdf.gz emd_33501_full_validation.pdf.gz | 846.2 KB | Display | |
| Data in XML |  emd_33501_validation.xml.gz emd_33501_validation.xml.gz | 21.3 KB | Display | |
| Data in CIF |  emd_33501_validation.cif.gz emd_33501_validation.cif.gz | 27.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33501 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33501 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33501 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33501 | HTTPS FTP |
-Related structure data
| Related structure data |  7xxfMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33501.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33501.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
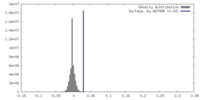
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map of odd number
| File | emd_33501_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map of odd number | ||||||||||||
| Projections & Slices |
| ||||||||||||
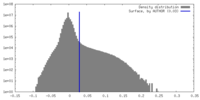
| Density Histograms |
-Half map: half map of even number
| File | emd_33501_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map of even number | ||||||||||||
| Projections & Slices |
| ||||||||||||
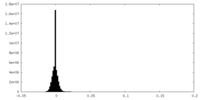
| Density Histograms |
- Sample components
Sample components
+Entire : Photosynthetic LH1-RC complex from the purple phototrophic bacter...
+Supramolecule #1: Photosynthetic LH1-RC complex from the purple phototrophic bacter...
+Macromolecule #1: Photosynthetic reaction center cytochrome c subunit
+Macromolecule #2: Reaction center protein L chain
+Macromolecule #3: Reaction center protein M chain
+Macromolecule #4: Photosynthetic reaction center H subunit
+Macromolecule #5: Light-harvesting protein
+Macromolecule #6: Light-harvesting protein
+Macromolecule #7: Light-harvesting protein LH1 Gamma-like
+Macromolecule #8: HEME C
+Macromolecule #9: (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-...
+Macromolecule #10: BACTERIOCHLOROPHYLL A
+Macromolecule #11: BACTERIOPHEOPHYTIN A
+Macromolecule #12: UBIQUINONE-10
+Macromolecule #13: DODECYL-BETA-D-MALTOSIDE
+Macromolecule #14: FE (III) ION
+Macromolecule #15: MENAQUINONE-9
+Macromolecule #16: (6~{E},8~{E},10~{E},12~{E},14~{E},16~{E},18~{E},20~{E},22~{E},24~...
+Macromolecule #17: CARDIOLIPIN
+Macromolecule #18: 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
+Macromolecule #19: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 277 K / Instrument: LEICA EM GP |
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average exposure time: 38.9 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.6 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)