[English] 日本語
 Yorodumi
Yorodumi- EMDB-33397: Hemichannel-focused structure of C-terminal truncated connexin43/... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (FIN conformation) | |||||||||
 Map data Map data | unsharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cx43 / Connexin / Gap junction channel / Gating mechanism / Membrane protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationgap junction channel activity involved in cardiac conduction electrical coupling / negative regulation of gonadotropin secretion / positive regulation of striated muscle tissue development / milk ejection reflex / positive regulation of morphogenesis of an epithelium / positive regulation of mesodermal cell differentiation / atrial ventricular junction remodeling / cell communication by chemical coupling / cell communication by electrical coupling / columnar/cuboidal epithelial cell maturation ...gap junction channel activity involved in cardiac conduction electrical coupling / negative regulation of gonadotropin secretion / positive regulation of striated muscle tissue development / milk ejection reflex / positive regulation of morphogenesis of an epithelium / positive regulation of mesodermal cell differentiation / atrial ventricular junction remodeling / cell communication by chemical coupling / cell communication by electrical coupling / columnar/cuboidal epithelial cell maturation / neuroblast migration / gap junction hemi-channel activity / negative regulation of trophoblast cell migration / monoatomic ion transmembrane transporter activity / microtubule-based transport / regulation of bone remodeling / regulation of ventricular cardiac muscle cell membrane depolarization / SARS-CoV-2 targets PDZ proteins in cell-cell junction / gap junction channel activity involved in cell communication by electrical coupling / contractile muscle fiber / glutathione transmembrane transporter activity / Oligomerization of connexins into connexons / Transport of connexins along the secretory pathway / Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane / gap junction assembly / atrial cardiac muscle cell action potential / cellular response to pH / Regulation of gap junction activity / cardiac conduction system development / regulation of atrial cardiac muscle cell membrane depolarization / Golgi-associated vesicle membrane / connexin complex / Formation of annular gap junctions / fascia adherens / Gap junction degradation / cell communication by electrical coupling involved in cardiac conduction / cell-cell contact zone / gap junction / bone remodeling / Gap junction assembly / skeletal muscle tissue regeneration / gap junction channel activity / export across plasma membrane / adult heart development / regulation of bone mineralization / regulation of ventricular cardiac muscle cell membrane repolarization / glutamate secretion / tight junction / blood vessel morphogenesis / lens development in camera-type eye / intermediate filament / xenobiotic transport / embryonic digit morphogenesis / maintenance of blood-brain barrier / positive regulation of stem cell proliferation / heart looping / RHOJ GTPase cycle / beta-tubulin binding / RHOQ GTPase cycle / efflux transmembrane transporter activity / establishment of mitotic spindle orientation / intercalated disc / lateral plasma membrane / alpha-tubulin binding / T cell proliferation / positive regulation of vascular associated smooth muscle cell proliferation / protein tyrosine kinase binding / tubulin binding / neuron migration / bone development / negative regulation of cell growth / beta-catenin binding / osteoblast differentiation / male gonad development / cellular response to amyloid-beta / protein localization / cell junction / cell-cell signaling / positive regulation of cold-induced thermogenesis / heart development / monoatomic ion transmembrane transport / scaffold protein binding / spermatogenesis / positive regulation of canonical NF-kappaB signal transduction / in utero embryonic development / membrane raft / apical plasma membrane / Golgi membrane / negative regulation of gene expression / intracellular membrane-bounded organelle / signaling receptor binding / focal adhesion / endoplasmic reticulum membrane / positive regulation of gene expression / Golgi apparatus / signal transduction / mitochondrion / nucleoplasm / nucleus / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
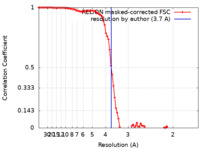
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Lee HJ / Cha HJ / Jeong H / Lee SN / Lee CW / Woo JS | |||||||||
| Funding support |  Korea, Republic Of, 1 items Korea, Republic Of, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM. Authors: Hyuk-Joon Lee / Hyung Jin Cha / Hyeongseop Jeong / Seu-Na Lee / Chang-Won Lee / Minsoo Kim / Jejoong Yoo / Jae-Sung Woo /  Abstract: Connexin family proteins assemble into hexameric hemichannels in the cell membrane. The hemichannels dock together between two adjacent membranes to form gap junction intercellular channels (GJIChs). ...Connexin family proteins assemble into hexameric hemichannels in the cell membrane. The hemichannels dock together between two adjacent membranes to form gap junction intercellular channels (GJIChs). We report the cryo-electron microscopy structures of Cx43 GJICh, revealing the dynamic equilibrium state of various channel conformations in detergents and lipid nanodiscs. We identify three different N-terminal helix conformations of Cx43-gate-covering (GCN), pore-lining (PLN), and flexible intermediate (FIN)-that are randomly distributed in purified GJICh particles. The conformational equilibrium shifts to GCN by cholesteryl hemisuccinates and to PLN by C-terminal truncations and at varying pH. While GJIChs that mainly comprise GCN protomers are occluded by lipids, those containing conformationally heterogeneous protomers show markedly different pore sizes. We observe an α-to-π-helix transition in the first transmembrane helix, which creates a side opening to the membrane in the FIN and PLN conformations. This study provides basic structural information to understand the mechanisms of action and regulation of Cx43 GJICh. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33397.map.gz emd_33397.map.gz | 48.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33397-v30.xml emd-33397-v30.xml emd-33397.xml emd-33397.xml | 13.8 KB 13.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33397_fsc.xml emd_33397_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_33397.png emd_33397.png | 8.8 KB | ||
| Filedesc metadata |  emd-33397.cif.gz emd-33397.cif.gz | 5.2 KB | ||
| Others |  emd_33397_half_map_1.map.gz emd_33397_half_map_1.map.gz emd_33397_half_map_2.map.gz emd_33397_half_map_2.map.gz | 48.6 MB 48.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33397 http://ftp.pdbj.org/pub/emdb/structures/EMD-33397 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33397 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33397 | HTTPS FTP |
-Validation report
| Summary document |  emd_33397_validation.pdf.gz emd_33397_validation.pdf.gz | 725.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33397_full_validation.pdf.gz emd_33397_full_validation.pdf.gz | 724.8 KB | Display | |
| Data in XML |  emd_33397_validation.xml.gz emd_33397_validation.xml.gz | 16.3 KB | Display | |
| Data in CIF |  emd_33397_validation.cif.gz emd_33397_validation.cif.gz | 21.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33397 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33397 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33397 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33397 | HTTPS FTP |
-Related structure data
| Related structure data |  7xqiMC  7f92C  7f93C  7f94C  7xq9C  7xqbC  7xqdC  7xqfC  7xqgC  7xqhC  7xqjC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33397.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33397.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.848 Å | ||||||||||||||||||||||||||||||||||||
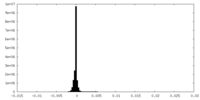
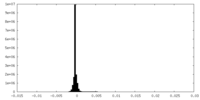
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map
| File | emd_33397_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
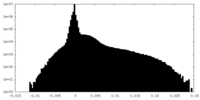
| Density Histograms |
-Half map: half map
| File | emd_33397_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
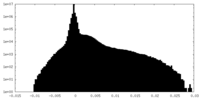
| Density Histograms |
- Sample components
Sample components
-Entire : Dodecameric complex of C-terminal deletion mutant of human Cx43/G...
| Entire | Name: Dodecameric complex of C-terminal deletion mutant of human Cx43/GJA1 (Cx43-M257) in lipid nanodiscs (POPE) |
|---|---|
| Components |
|
-Supramolecule #1: Dodecameric complex of C-terminal deletion mutant of human Cx43/G...
| Supramolecule | Name: Dodecameric complex of C-terminal deletion mutant of human Cx43/GJA1 (Cx43-M257) in lipid nanodiscs (POPE) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Hemichannel of Human Cx43-M257 gap junction channel with flexible-intermediate NTH conformation |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Gap junction alpha-1 protein
| Macromolecule | Name: Gap junction alpha-1 protein / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 29.396547 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGDWSALGKL LDKVQAYSTA GGKVWLSVLF IFRILLLGTA VESAWGDEQS AFRCNTQQPG CENVCYDKSF PISHVRFWVL QIIFVSVPT LLYLAHVFYV MRKEEKLNKK EEELKVAQTD GVNVDMHLKQ IEIKKFKYGI EEHGKVKMRG GLLRTYIISI L FKSIFEVA ...String: MGDWSALGKL LDKVQAYSTA GGKVWLSVLF IFRILLLGTA VESAWGDEQS AFRCNTQQPG CENVCYDKSF PISHVRFWVL QIIFVSVPT LLYLAHVFYV MRKEEKLNKK EEELKVAQTD GVNVDMHLKQ IEIKKFKYGI EEHGKVKMRG GLLRTYIISI L FKSIFEVA FLLIQWYIYG FSLSAVYTCK RDPCPHQVDC FLSRPTEKTI FIIFMLVVSL VSLALNIIEL FYVFFKGVKD RV KGKSDPY HATSGALSPA UniProtKB: Gap junction alpha-1 protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 72.42 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.9 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)