[English] 日本語
 Yorodumi
Yorodumi- EMDB-32377: 2.02 angstrom cryo-EM structure of the pump-like channelrhodopsin... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-32377 | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | 2.02 angstrom cryo-EM structure of the pump-like channelrhodopsin ChRmine | |||||||||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||
 Keywords Keywords | channelrhodopsin / ion channel / photoreceptor / cryo-EM / MEMBRANE PROTEIN | |||||||||||||||||||||||||||
| Biological species |  Rhodomonas lens (eukaryote) Rhodomonas lens (eukaryote) | |||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.02 Å | |||||||||||||||||||||||||||
 Authors Authors | Kishi KE / Kim Y / Fukuda M / Yamashita K / Deisseroth K / Kato HE | |||||||||||||||||||||||||||
| Funding support |  Japan, 8 items Japan, 8 items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2022 Journal: Cell / Year: 2022Title: Structural basis for channel conduction in the pump-like channelrhodopsin ChRmine. Authors: Koichiro E Kishi / Yoon Seok Kim / Masahiro Fukuda / Masatoshi Inoue / Tsukasa Kusakizako / Peter Y Wang / Charu Ramakrishnan / Eamon F X Byrne / Elina Thadhani / Joseph M Paggi / Toshiki E ...Authors: Koichiro E Kishi / Yoon Seok Kim / Masahiro Fukuda / Masatoshi Inoue / Tsukasa Kusakizako / Peter Y Wang / Charu Ramakrishnan / Eamon F X Byrne / Elina Thadhani / Joseph M Paggi / Toshiki E Matsui / Keitaro Yamashita / Takashi Nagata / Masae Konno / Sean Quirin / Maisie Lo / Tyler Benster / Tomoko Uemura / Kehong Liu / Mikihiro Shibata / Norimichi Nomura / So Iwata / Osamu Nureki / Ron O Dror / Keiichi Inoue / Karl Deisseroth / Hideaki E Kato /    Abstract: ChRmine, a recently discovered pump-like cation-conducting channelrhodopsin, exhibits puzzling properties (large photocurrents, red-shifted spectrum, and extreme light sensitivity) that have created ...ChRmine, a recently discovered pump-like cation-conducting channelrhodopsin, exhibits puzzling properties (large photocurrents, red-shifted spectrum, and extreme light sensitivity) that have created new opportunities in optogenetics. ChRmine and its homologs function as ion channels but, by primary sequence, more closely resemble ion pump rhodopsins; mechanisms for passive channel conduction in this family have remained mysterious. Here, we present the 2.0 Å resolution cryo-EM structure of ChRmine, revealing architectural features atypical for channelrhodopsins: trimeric assembly, a short transmembrane-helix 3, a twisting extracellular-loop 1, large vestibules within the monomer, and an opening at the trimer interface. We applied this structure to design three proteins (rsChRmine and hsChRmine, conferring further red-shifted and high-speed properties, respectively, and frChRmine, combining faster and more red-shifted performance) suitable for fundamental neuroscience opportunities. These results illuminate the conduction and gating of pump-like channelrhodopsins and point the way toward further structure-guided creation of channelrhodopsins for applications across biology. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32377.map.gz emd_32377.map.gz | 2.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32377-v30.xml emd-32377-v30.xml emd-32377.xml emd-32377.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32377_fsc.xml emd_32377_fsc.xml | 15.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_32377.png emd_32377.png | 165.2 KB | ||
| Masks |  emd_32377_msk_1.map emd_32377_msk_1.map | 16.2 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-32377.cif.gz emd-32377.cif.gz | 5.9 KB | ||
| Others |  emd_32377_half_map_1.map.gz emd_32377_half_map_1.map.gz emd_32377_half_map_2.map.gz emd_32377_half_map_2.map.gz | 14.9 MB 14.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32377 http://ftp.pdbj.org/pub/emdb/structures/EMD-32377 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32377 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32377 | HTTPS FTP |
-Related structure data
| Related structure data |  7w9wMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10926 (Title: The pump-like chanelrhodopsin ChRmine / Data size: 850.8 EMPIAR-10926 (Title: The pump-like chanelrhodopsin ChRmine / Data size: 850.8 Data #1: Multiframe micrographs for Structural basis for channel conduction in the pump-like channelrhodopsin ChRmine [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32377.map.gz / Format: CCP4 / Size: 16.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32377.map.gz / Format: CCP4 / Size: 16.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.94318 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
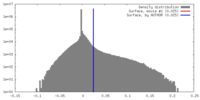
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
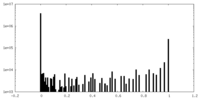
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_32377_msk_1.map emd_32377_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
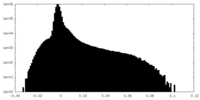
| Density Histograms |
-Half map: #1
| File | emd_32377_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
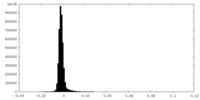
| Density Histograms |
-Half map: #2
| File | emd_32377_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ChRmine
| Entire | Name: ChRmine |
|---|---|
| Components |
|
-Supramolecule #1: ChRmine
| Supramolecule | Name: ChRmine / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Rhodomonas lens (eukaryote) Rhodomonas lens (eukaryote) |
-Macromolecule #1: ChRmine
| Macromolecule | Name: ChRmine / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Rhodomonas lens (eukaryote) Rhodomonas lens (eukaryote) |
| Molecular weight | Theoretical: 35.772965 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPMAHAPGTD QMFYVGTMDG WYLDTKLNSV AIGAHWSCFI VLTITTFYLG YESWTSRGPS KRTSFYAGYQ EEQNLALFVN FFAMLSYFG KIVADTLGHN FGDVGPFIIG FGNYRYADYM LTCPMLVYDL LYQLRAPYRV SCSAIIFAIL MSGVLAEFYA E GDPRLRNG ...String: GPMAHAPGTD QMFYVGTMDG WYLDTKLNSV AIGAHWSCFI VLTITTFYLG YESWTSRGPS KRTSFYAGYQ EEQNLALFVN FFAMLSYFG KIVADTLGHN FGDVGPFIIG FGNYRYADYM LTCPMLVYDL LYQLRAPYRV SCSAIIFAIL MSGVLAEFYA E GDPRLRNG AYAWYGFGCF WFIFAYSIVM SIVAKQYSRL AQLAQDTGAE HSLHVLKFAV FTFSMLWILF PLVWAICPRG FG WIDDNWT EVAHCVCDIV AKSCYGFALA RFRKTYDEEL FRLLEQLGHD EDEFQKLELD MRLSSNGERL EVLFQ |
-Macromolecule #2: RETINAL
| Macromolecule | Name: RETINAL / type: ligand / ID: 2 / Number of copies: 1 / Formula: RET |
|---|---|
| Molecular weight | Theoretical: 284.436 Da |
| Chemical component information |  ChemComp-RET: |
-Macromolecule #3: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 3 / Number of copies: 2 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #4: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 4 / Number of copies: 5 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 48 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: GOLD / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 51.533 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)