[English] 日本語
 Yorodumi
Yorodumi- EMDB-31944: Pentacylindrical allophycocyanin core from Thermosynechococcus vu... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Pentacylindrical allophycocyanin core from Thermosynechococcus vulcanus | ||||||||||||
 Map data Map data | Cryo-EM map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Light-harvesting complex / Phycobilisome / Energy transfer / PHOTOSYNTHESIS | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationphycobilisome / plasma membrane-derived thylakoid membrane / photosynthesis / lyase activity Similarity search - Function | ||||||||||||
| Biological species |  Thermosynechococcus vulcanus NIES-2134 (bacteria) / Thermosynechococcus vulcanus NIES-2134 (bacteria) /   Thermosynechococcus vestitus BP-1 (bacteria) Thermosynechococcus vestitus BP-1 (bacteria) | ||||||||||||
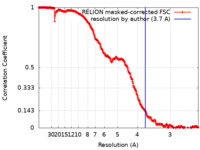
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | ||||||||||||
 Authors Authors | Kawakami K / Hamaguchi T / Hirose Y / Kosumi D / Miyata M / Kamiya N / Yonekura K | ||||||||||||
| Funding support |  Japan, 3 items Japan, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Core and rod structures of a thermophilic cyanobacterial light-harvesting phycobilisome. Authors: Keisuke Kawakami / Tasuku Hamaguchi / Yuu Hirose / Daisuke Kosumi / Makoto Miyata / Nobuo Kamiya / Koji Yonekura /  Abstract: Cyanobacteria, glaucophytes, and rhodophytes utilize giant, light-harvesting phycobilisomes (PBSs) for capturing solar energy and conveying it to photosynthetic reaction centers. PBSs are ...Cyanobacteria, glaucophytes, and rhodophytes utilize giant, light-harvesting phycobilisomes (PBSs) for capturing solar energy and conveying it to photosynthetic reaction centers. PBSs are compositionally and structurally diverse, and exceedingly complex, all of which pose a challenge for a comprehensive understanding of their function. To date, three detailed architectures of PBSs by cryo-electron microscopy (cryo-EM) have been described: a hemiellipsoidal type, a block-type from rhodophytes, and a cyanobacterial hemidiscoidal-type. Here, we report cryo-EM structures of a pentacylindrical allophycocyanin core and phycocyanin-containing rod of a thermophilic cyanobacterial hemidiscoidal PBS. The structures define the spatial arrangement of protein subunits and chromophores, crucial for deciphering the energy transfer mechanism. They reveal how the pentacylindrical core is formed, identify key interactions between linker proteins and the bilin chromophores, and indicate pathways for unidirectional energy transfer. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31944.map.gz emd_31944.map.gz | 94.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31944-v30.xml emd-31944-v30.xml emd-31944.xml emd-31944.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31944_fsc.xml emd_31944_fsc.xml | 28.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_31944.png emd_31944.png | 100.5 KB | ||
| Filedesc metadata |  emd-31944.cif.gz emd-31944.cif.gz | 7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31944 http://ftp.pdbj.org/pub/emdb/structures/EMD-31944 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31944 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31944 | HTTPS FTP |
-Related structure data
| Related structure data |  7veaMC  7vebC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_31944.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31944.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.24 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Phycobilisome core complex from Thermosynechococcus vulcanus
| Entire | Name: Phycobilisome core complex from Thermosynechococcus vulcanus |
|---|---|
| Components |
|
-Supramolecule #1: Phycobilisome core complex from Thermosynechococcus vulcanus
| Supramolecule | Name: Phycobilisome core complex from Thermosynechococcus vulcanus type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Thermosynechococcus vulcanus NIES-2134 (bacteria) Thermosynechococcus vulcanus NIES-2134 (bacteria) |
-Macromolecule #1: Allophycocyanin alpha chain
| Macromolecule | Name: Allophycocyanin alpha chain / type: protein_or_peptide / ID: 1 / Number of copies: 40 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermosynechococcus vestitus BP-1 (bacteria) / Strain: BP-1 Thermosynechococcus vestitus BP-1 (bacteria) / Strain: BP-1 |
| Molecular weight | Theoretical: 17.555926 KDa |
| Sequence | String: MSVVTKSIVN ADAEARYLSP GELDRIKNFV STGERRLRIA QTLTENRERI VKQAGDQLFQ KRPDVVSPGG NAYGEEMTAT CLRDLDYYL RLVTYGIVAG DVTPIEEIGL VGVREMYNSL GTPIPAVAEG IRAMKNVACS LLSAEDAAEA GSYFDFVIGA M Q UniProtKB: Allophycocyanin alpha chain |
-Macromolecule #2: Allophycocyanin beta chain
| Macromolecule | Name: Allophycocyanin beta chain / type: protein_or_peptide / ID: 2 / Number of copies: 40 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermosynechococcus vestitus BP-1 (bacteria) / Strain: BP-1 Thermosynechococcus vestitus BP-1 (bacteria) / Strain: BP-1 |
| Molecular weight | Theoretical: 17.388877 KDa |
| Sequence | String: MQDAITAVIN ASDVQGKYLD TAAMEKLKAY FATGELRVRA ASVISANAAN IVKEAVAKSL LYSDITRPGG (MEN)MYTTR RYA ACIRDLDYYL RYATYAMLAG DPSILDERVL NGLKETYNSL GVPIAATVQA IQAMKEVTAS LVGADAGKEM GIYFDYI CS GLS UniProtKB: Allophycocyanin beta chain |
-Macromolecule #3: Phycobiliprotein ApcE
| Macromolecule | Name: Phycobiliprotein ApcE / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermosynechococcus vestitus BP-1 (bacteria) / Strain: BP-1 Thermosynechococcus vestitus BP-1 (bacteria) / Strain: BP-1 |
| Molecular weight | Theoretical: 127.772922 KDa |
| Sequence | String: MVVKASGGSS VARPQLYQTV PVSTIIQAEQ QDRFLNRGEL DELAVYLRSG AKRLEIATTL TRNADIIVSR AANRIFVGGS PMAFLSRPQ TEEAPQFTTG ARGEAIDIKE AMKLGTATYV DTRGGFLEGL RSIFSASSGG APVGFKPINI ARYGPARMEK S LRDLDWFL ...String: MVVKASGGSS VARPQLYQTV PVSTIIQAEQ QDRFLNRGEL DELAVYLRSG AKRLEIATTL TRNADIIVSR AANRIFVGGS PMAFLSRPQ TEEAPQFTTG ARGEAIDIKE AMKLGTATYV DTRGGFLEGL RSIFSASSGG APVGFKPINI ARYGPARMEK S LRDLDWFL RYTTYAIVAG DPNILAVNTR GLREIIEAAC SSDATIAALQ EMRRAALSYF EKDAEAKGIV ETYFDVLINE FI APAPSDK VRQRNSTDLQ GLQLPQIYFN AAERRPKFVM KPGLSAAEKN EVVKAAYRQI FERDISRAYG LGISDLESKV KNG SISMKE FIRQLAKSPL YRKNFYEPYI NSRALELAFR HILGRGPSSR EEVQTYFAII SKGGLPALVD ALVDSKEYSD YFGE ETVPY LRGLGQEAQE CRNWGAQQDL FKYSAPFRKV PQFITTFAAQ DQPLPDQHPY GSGNDPLEIQ FGAIFPKEKK NPSAR PQPF NKDTRRILIA RGPGINNQVS NPGARGLTPG TLGPKVFKLD QLPSINARIG KRSIATGTDS VKFAESSTQR VIRAAY LQV FGRDVYEGQR QKVAEIKLEN GEISVREFVR ILAKSNLFRS LYWTPLYVTK AIEYIHRRLL GRPTYGRQEM NAYFDIA SK KGLYGLVDAI IDSQEYSEAF GEDTVPYERY ITPQGLALRS LRVGTIGETG VPPEKEETPR FVELGAVTEL RTEPAIQF R ANQGVSKRRE QTKVFKLTDL NDKQNLQLVI QAAYRQVFER DVAPYIVRDE FTALESKLSN GEITLKEFIE ALGCSELYQ KEFYTPYPNT KVIELGTKHF LGRAPLDQAE IRRYNQILAT QGLKAFVQAL VSSAEYAQAF GEDTVPYRRF PTLPAANFPN TEKLHNQLT KQSDAIVVPS FAPVKPRLDN TKLPLLSRAI AEQEAKARQA DPSKPRFIEL GRSFRNGDGQ SVEVGVGTTR R RPARIFRM TVGAPSAEVE LVINAIYCQV MDVFSGQVPS QFRRPDLESR LRNGEITVRE FVRTLASSEI YRNRFYTPYP NT KVIEFLF RHLLGRAPAT QAEIRQYNKI LADQGLKTAV ETMVNSPEYS RYFGEDVVPY KRFPTLPAGN YIGSVKADAD LVK QSWSSL SPSLVGIQPS HRD UniProtKB: Phycobiliprotein ApcE |
-Macromolecule #4: Phycobilisome core component
| Macromolecule | Name: Phycobilisome core component / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermosynechococcus vestitus BP-1 (bacteria) / Strain: BP-1 Thermosynechococcus vestitus BP-1 (bacteria) / Strain: BP-1 |
| Molecular weight | Theoretical: 18.757221 KDa |
| Sequence | String: MRDAVTTLIK NYDSTGRYLD RDAVDRLRSY FNSGAARVKA AAVINANAAA IVKEAASALF TEQPELIQPG G(MEN)AYTT RRY ATCLRDMDYY LRYASYAIVA GDVDVLNERV LEGLRETYNS LGVPIGPTVR GIQIMKEIVR DRVAAAGIED TGIVEQP FD YMCRQLSEVN I UniProtKB: Phycobilisome core component |
-Macromolecule #5: Phycobilisome 7.8 kDa linker polypeptide, allophycocyanin-associa...
| Macromolecule | Name: Phycobilisome 7.8 kDa linker polypeptide, allophycocyanin-associated, core type: protein_or_peptide / ID: 5 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermosynechococcus vestitus BP-1 (bacteria) / Strain: BP-1 Thermosynechococcus vestitus BP-1 (bacteria) / Strain: BP-1 |
| Molecular weight | Theoretical: 7.876255 KDa |
| Sequence | String: MRMFKITACV PSQTRIRTQR ELQNTYFTKL VPYENWFREQ QRIQKMGGKI VKVELFTGKP GVNTGLA UniProtKB: Phycobilisome 7.8 kDa linker polypeptide, allophycocyanin-associated, core |
-Macromolecule #6: PHYCOCYANOBILIN
| Macromolecule | Name: PHYCOCYANOBILIN / type: ligand / ID: 6 / Number of copies: 84 / Formula: CYC |
|---|---|
| Molecular weight | Theoretical: 588.694 Da |
| Chemical component information |  ChemComp-CYC: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.5 |
|---|---|
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 84.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 150.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)