+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Phycocyanin rod structure of cyanobacterial phycobilisome | ||||||||||||
 Map data Map data | postprocess_masked.mrc | ||||||||||||
 Sample Sample |
| ||||||||||||
| Function / homology |  Function and homology information Function and homology informationphycobilisome / plasma membrane-derived thylakoid membrane / photosynthesis Similarity search - Function | ||||||||||||
| Biological species |  Thermosynechococcus vulcanus NIES-2134 (bacteria) / Thermosynechococcus vulcanus NIES-2134 (bacteria) /   Thermosynechococcus elongatus BP-1 (bacteria) Thermosynechococcus elongatus BP-1 (bacteria) | ||||||||||||
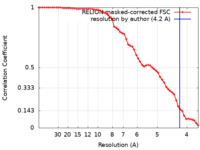
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | ||||||||||||
 Authors Authors | Kawakami K / Hamaguchi T / Hirose Y / Kosumi D / Miyata M / Kamiya N / Yonekura K | ||||||||||||
| Funding support |  Japan, 3 items Japan, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Core and rod structures of a thermophilic cyanobacterial light-harvesting phycobilisome. Authors: Keisuke Kawakami / Tasuku Hamaguchi / Yuu Hirose / Daisuke Kosumi / Makoto Miyata / Nobuo Kamiya / Koji Yonekura /  Abstract: Cyanobacteria, glaucophytes, and rhodophytes utilize giant, light-harvesting phycobilisomes (PBSs) for capturing solar energy and conveying it to photosynthetic reaction centers. PBSs are ...Cyanobacteria, glaucophytes, and rhodophytes utilize giant, light-harvesting phycobilisomes (PBSs) for capturing solar energy and conveying it to photosynthetic reaction centers. PBSs are compositionally and structurally diverse, and exceedingly complex, all of which pose a challenge for a comprehensive understanding of their function. To date, three detailed architectures of PBSs by cryo-electron microscopy (cryo-EM) have been described: a hemiellipsoidal type, a block-type from rhodophytes, and a cyanobacterial hemidiscoidal-type. Here, we report cryo-EM structures of a pentacylindrical allophycocyanin core and phycocyanin-containing rod of a thermophilic cyanobacterial hemidiscoidal PBS. The structures define the spatial arrangement of protein subunits and chromophores, crucial for deciphering the energy transfer mechanism. They reveal how the pentacylindrical core is formed, identify key interactions between linker proteins and the bilin chromophores, and indicate pathways for unidirectional energy transfer. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31945.map.gz emd_31945.map.gz | 1.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31945-v30.xml emd-31945-v30.xml emd-31945.xml emd-31945.xml | 14.3 KB 14.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31945_fsc.xml emd_31945_fsc.xml | 5.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_31945.png emd_31945.png | 65.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31945 http://ftp.pdbj.org/pub/emdb/structures/EMD-31945 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31945 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31945 | HTTPS FTP |
-Validation report
| Summary document |  emd_31945_validation.pdf.gz emd_31945_validation.pdf.gz | 381.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31945_full_validation.pdf.gz emd_31945_full_validation.pdf.gz | 380.8 KB | Display | |
| Data in XML |  emd_31945_validation.xml.gz emd_31945_validation.xml.gz | 8.6 KB | Display | |
| Data in CIF |  emd_31945_validation.cif.gz emd_31945_validation.cif.gz | 10.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31945 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31945 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31945 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31945 | HTTPS FTP |
-Related structure data
| Related structure data |  7vebMC  7veaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_31945.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31945.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | postprocess_masked.mrc | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Phycocyanin rod complex of cyanobacterial phyocobilisome
| Entire | Name: Phycocyanin rod complex of cyanobacterial phyocobilisome |
|---|---|
| Components |
|
-Supramolecule #1: Phycocyanin rod complex of cyanobacterial phyocobilisome
| Supramolecule | Name: Phycocyanin rod complex of cyanobacterial phyocobilisome type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Thermosynechococcus vulcanus NIES-2134 (bacteria) Thermosynechococcus vulcanus NIES-2134 (bacteria) |
-Macromolecule #1: C-phycocyanin alpha subunit
| Macromolecule | Name: C-phycocyanin alpha subunit / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermosynechococcus elongatus BP-1 (bacteria) / Strain: BP-1 Thermosynechococcus elongatus BP-1 (bacteria) / Strain: BP-1 |
| Molecular weight | Theoretical: 17.456631 KDa |
| Sequence | String: MKTPITEAIA AADTQGRFLS NTELQAVDGR FKRAVASMEA ARALTNNAQS LIDGAAQAVY QKFPYTTTMQ GSQYASTPEG KAKCARDIG YYLRMVTYCL VAGGTGPMDE YLIAGLSEIN STFDLSPSWY IEALKYIKAN HGLTGQAAVE ANAYIDYAIN A LS |
-Macromolecule #2: C-phycocyanin beta subunit
| Macromolecule | Name: C-phycocyanin beta subunit / type: protein_or_peptide / ID: 2 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermosynechococcus elongatus BP-1 (bacteria) / Strain: BP-1 Thermosynechococcus elongatus BP-1 (bacteria) / Strain: BP-1 |
| Molecular weight | Theoretical: 18.216652 KDa |
| Sequence | String: MLDAFAKVVA QADARGEFLT NAQFDALSNL VKEGNKRLDA VNRITSNAST IVANAARALF AEQPQLIQPG G(MEN)AYTN RRM AACLRDMEII LRYVTYAILA GDSSVLDDRC LNGLRETYQA LGTPGSSVAV AIQKMKDAAI AIANDPNGIT PGDCSAL MS EIAGYFDRAA AAVA |
-Macromolecule #3: Phycobilisome 8.9 kDa linker polypeptide, phycocyanin-associated, rod
| Macromolecule | Name: Phycobilisome 8.9 kDa linker polypeptide, phycocyanin-associated, rod type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermosynechococcus elongatus BP-1 (bacteria) / Strain: BP-1 Thermosynechococcus elongatus BP-1 (bacteria) / Strain: BP-1 |
| Molecular weight | Theoretical: 8.681903 KDa |
| Sequence | String: MFGQTASGSA ALSPSGARVF RYEVVGLRQN EETDRMEFPI RRSGSTFITV PYNRMNEEMQ RITRMGGKIV SITPVVAS |
-Macromolecule #4: Phycobilisome 32.1 kDa linker polypeptide, phycocyanin-associated, rod
| Macromolecule | Name: Phycobilisome 32.1 kDa linker polypeptide, phycocyanin-associated, rod type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermosynechococcus elongatus BP-1 (bacteria) / Strain: BP-1 Thermosynechococcus elongatus BP-1 (bacteria) / Strain: BP-1 |
| Molecular weight | Theoretical: 32.158914 KDa |
| Sequence | String: MAITAAASRL GTSAFSDAPP VELRANWSEE DLETVIRAVY RQVLGNDYVM ASERLVSAES LLRNGKITVR EFVRAVAKSE LYKEKFLYG NFQTRVIELN YKHLLGRAPY DESEVIFHLD LYENEGFDAD IDSYIDSPEY TNSFGDWVVP YYRGFNTQPG Q KTVGFNRI ...String: MAITAAASRL GTSAFSDAPP VELRANWSEE DLETVIRAVY RQVLGNDYVM ASERLVSAES LLRNGKITVR EFVRAVAKSE LYKEKFLYG NFQTRVIELN YKHLLGRAPY DESEVIFHLD LYENEGFDAD IDSYIDSPEY TNSFGDWVVP YYRGFNTQPG Q KTVGFNRI FRLYRGYANS DRAQAEGSMS RLARDLATNR ANTVVPPSNS DTAFAYYTPS ADVPPRACLG GSFGESGRVY RI EVAGIRQ PGYPGVRRSS TAFLVPYEQL SAKMQQLQRT GARIISVNPA |
-Macromolecule #5: Phycobilisome rod-core linker polypeptide CpcG2
| Macromolecule | Name: Phycobilisome rod-core linker polypeptide CpcG2 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermosynechococcus elongatus BP-1 (bacteria) / Strain: BP-1 Thermosynechococcus elongatus BP-1 (bacteria) / Strain: BP-1 |
| Molecular weight | Theoretical: 28.817357 KDa |
| Sequence | String: MTIPLLSYAP SSQNQRVAGY EVPNEETPWR YSLEDAVDQS DIDELIWAAY RQVFSEHVVL KSTRQPHLES QLANRAISVR DFIRGLAKS ETFRRLVVET NSNYRLVEIA LKRLLGRAPY NKQEELAWSI RIATDGWQKF VDTLVDSDEY TQNFGDNTVP Y QRRRYKDR ...String: MTIPLLSYAP SSQNQRVAGY EVPNEETPWR YSLEDAVDQS DIDELIWAAY RQVFSEHVVL KSTRQPHLES QLANRAISVR DFIRGLAKS ETFRRLVVET NSNYRLVEIA LKRLLGRAPY NKQEELAWSI RIATDGWQKF VDTLVDSDEY TQNFGDNTVP Y QRRRYKDR PFNLVTPRYS DYWRDKLENS RYKWGDIRNF LEMARSVKVT PVQFKPVSTA NVQIPDTTRR DRPTVPASIN PT ASFPLR |
-Macromolecule #6: PHYCOCYANOBILIN
| Macromolecule | Name: PHYCOCYANOBILIN / type: ligand / ID: 6 / Number of copies: 36 / Formula: CYC |
|---|---|
| Molecular weight | Theoretical: 588.694 Da |
| Chemical component information |  ChemComp-CYC: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.5 |
|---|---|
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 150.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)